Method, device, equipment and storage medium for predicting protein binding sites
A technology for combining sites and prediction methods, applied in the field of bioinformatics, which can solve the problems of low accuracy and generality
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
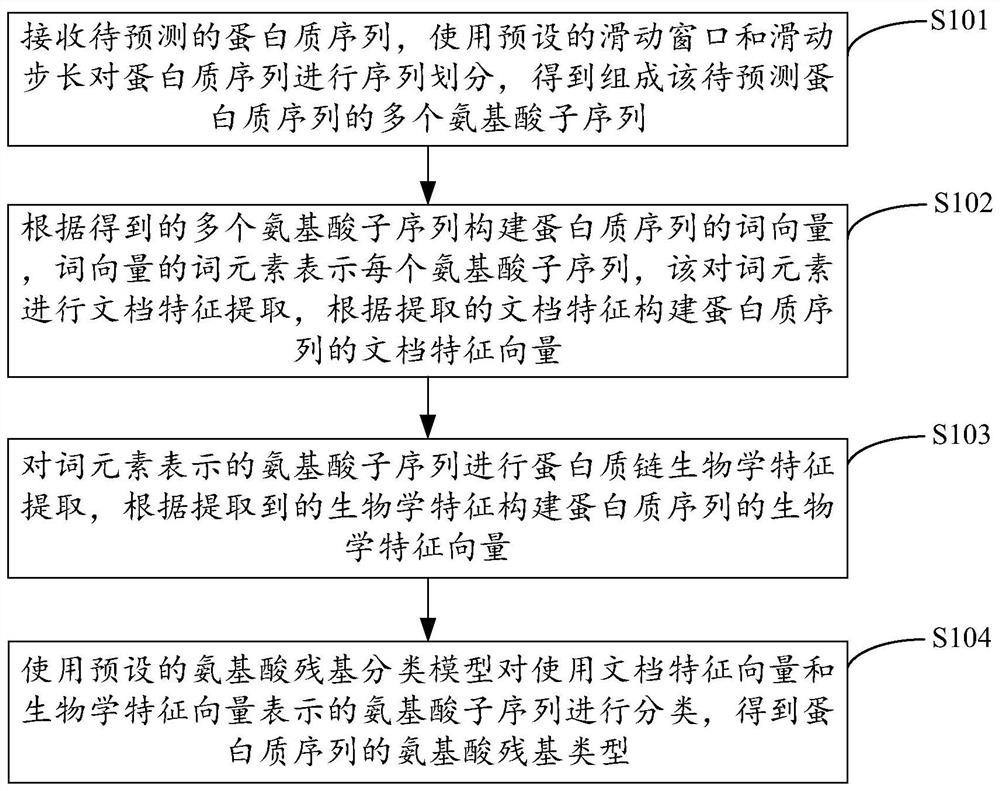
[0026] figure 1 The implementation process of the method for predicting protein binding sites provided by Example 1 of the present invention is shown. For the convenience of illustration, only the parts related to the embodiment of the present invention are shown, and the details are as follows:
[0027] In step S101, the protein sequence to be predicted is received, and the protein sequence is segmented using a preset sliding window and sliding step to obtain a plurality of amino acid subsequences constituting the protein sequence to be predicted.
[0028] The embodiment of the present invention is applicable to the prediction system of protein binding sites. In the embodiment of the present invention, in order to reflect the aggregation characteristics of protein-protein binding sites, after receiving the protein sequence to be predicted, the sliding window is started, and the protein sequence is divided by adjusting the size of the sliding window and the sliding step length...
Embodiment 2
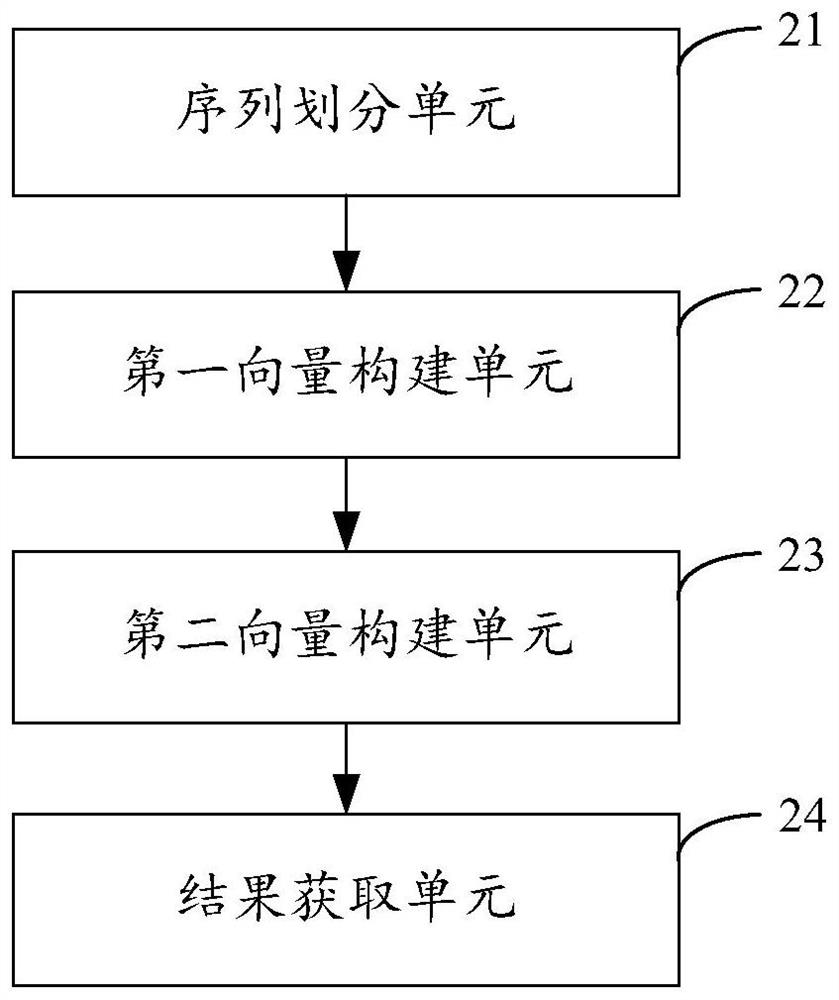
[0041] figure 2 The structure of the protein binding site prediction device provided in Example 2 of the present invention is shown. For the convenience of description, only the parts related to the embodiment of the present invention are shown, including:
[0042] The sequence division unit 21 is configured to receive the protein sequence to be predicted, and use a preset sliding window and sliding step to perform sequence division on the protein sequence, to obtain multiple amino acid subsequences constituting the protein sequence to be predicted.
[0043] The first vector construction unit 22 is used to construct the word vector of the protein sequence according to the obtained multiple amino acid subsequences, the word element of the word vector represents each amino acid subsequence, and extracts the document features of the word elements, and constructs according to the extracted document features Document feature vectors for protein sequences.
[0044] The second vect...
Embodiment 3
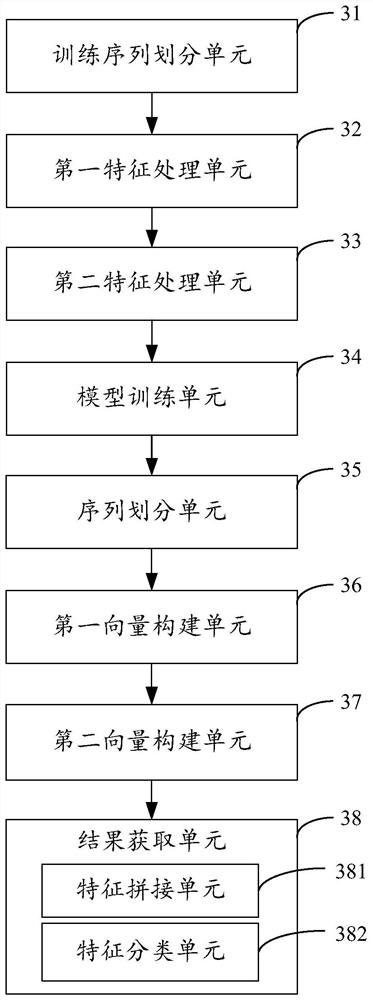
[0049] image 3 The structure of the protein binding site prediction device provided by Example 3 of the present invention is shown. For the convenience of description, only the parts related to the embodiment of the present invention are shown, including:
[0050] The training sequence division unit 31 is configured to perform sequence division on the training protein sequence in the preset training set by using a preset sliding window and a sliding step to obtain a plurality of training amino acid subsequences constituting the training protein sequence.
[0051] The first feature processing unit 32 is used to construct the training word vector of the training protein sequence according to the obtained multiple training amino acid subsequences, the training word element of the training word vector represents each training amino acid subsequence, and performs document feature extraction on the training word element , construct a document feature training vector for training pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com