A kind of identification method of bletilla striata
A technology of Bletilla striata and Bletilla striata, which is applied to biochemical equipment and methods, measurement/inspection of microorganisms, etc., can solve problems such as low drying rate, wild resources of Bletilla striata that cannot meet market demand, and difficulty in distinguishing Bletilla striata purple or Bletilla striata yellow, etc. To achieve the effect of effective identification method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] A method for identifying Bletilla striata, comprising the following steps:
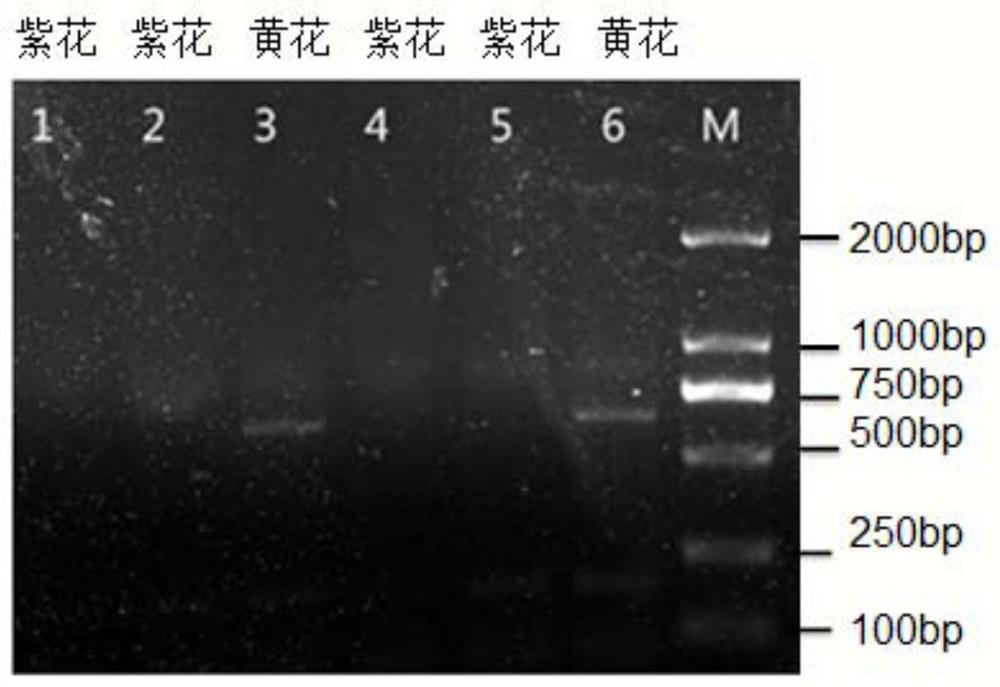
[0023] a. Select 3 Bletilla striata strains and 3 Bletilla striata strains with the same year, same growth conditions and same plant size, and use the CTAB method to extract individual genomic DNA respectively; then use amplification primers DUAN5: 5'-TAGTAACGGCGAGCGAAC-3', DUNA65 '-GGCATAGTTCACCATCTTTC-3' was amplified by PCR, and the size of the amplified product was 760bp; the amplification conditions were pre-denaturation at 94°C for 5 min, denaturation at 94°C for 1 min, annealing at 58°C for 45 s, and extension at 72°C for 1 min , according to this cycle 30 times, and finally extended at a temperature of 72°C for 10 min, and stored at a temperature of 4°C; the initial reaction system for PCR amplification is: 30 μL system containing 6 μL of 5×Taq buffer, 2.4 μL of dNTPs, 1 μL of each primer, and 1 μL of primers. Final concentration 0.4μM, template 1μL, template final concentration about 6...
Embodiment 2
[0028] A method for identifying Bletilla striata, comprising the following steps:
[0029] a. Select 60 Bletilla striata individuals with the same planting time, including 2 Bletilla striata chrysalis;
[0030] b. Using the CTAB method to extract 60 individual genomic DNAs respectively;
[0031] c. Use amplification primers DUAN5: 5'-TAGTAACGGCGAGCGAAC-3' and JDHH: 5'-TTATCAGCTTCCTTGCGCCCG-3' for PCR amplification; PCR amplification conditions are: pre-denaturation temperature 94°C, time 5min; denaturation temperature 94°C, Time 1min; annealing temperature 54°C, time 45s; extension temperature 72°C, time 1min; press this cycle 30 times, finally extend at 72°C for 10 minutes, store in refrigerator at 4°C; PCR amplification system is 30μL system: which contains 5×Taqbuffer 6μL, dNTPs 2.4μL, upstream and downstream primers 1μL each, primer final concentration 0.4μM, template 1μL, template final concentration about 60ng, Taq enzyme 0.15μL, add ddH 2 0 to 30 μL;
[0032] d. Perf...
Embodiment 3
[0034] A method for identifying Bletilla striata, comprising the following steps:
[0035] a. Select 100 Bletilla striata individuals with the same planting time, including 5 Bletilla striata chrysalis;
[0036] b. Using the CTAB method to extract 100 individual genomic DNAs respectively;
[0037]c. Use amplification primers DUAN5: 5'-TAGTAACGGCGAGCGAAC-3' and JDHH: 5'-TTATCAGCTTCCTTGCGCCCG-3' for PCR amplification; PCR amplification conditions are: pre-denaturation temperature 94°C, time 5min; denaturation temperature 94°C, Time 1min; annealing temperature 54°C, time 45s; extension temperature 72°C, time 1min; press this cycle 30 times, finally extend at 72°C for 10 minutes, store in refrigerator at 4°C; PCR amplification system is 30μL system: which contains 5×Taqbuffer 6μL, dNTPs 2.4μL, upstream and downstream primers 1μL each, primer final concentration 0.4μM, template 1μL, template final concentration about 60ng, Taq enzyme 0.15μL, add ddH 2 0 to 30 μL;
[0038] d. Car...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com