Identification method of ST37 type clostridium difficile and unit point variation type ST81 thereof
A Clostridium difficile and identification method technology, which is applied in the field of strain type identification, can solve the problems of cumbersome operation of the identification method, inability to use conventional detection, and long time-consuming, etc., and achieves easy promotion and application, simple method, and easy operation Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Samples: 159 strains of Clostridium difficile were obtained from the Hebei Medical Microbial Culture Collection Center (HBCMCC), among which 152 strains of Clostridium difficile were randomly selected from diarrhea patients and healthy population isolates collected from 2010 to 2014 by using the method of random number table (65 strains were collected from patients with diarrhea, 87 strains were collected from healthy adults or children), and 7 strains of Clostridium difficile standard strains (ATCC BAA-1870, ATCC BAA-1382, ATCC BAA-1812, ATCC BAA-1812, ATCC BAA-1804, ATCC9689, ATCC 43255 and ATCC 43598).
[0023] The strains were stored in the HBCMCC ultra-low temperature refrigerator, and the strains were resuscitated before use. The strains were inoculated on Columbia blood agar plates (OXOID, UK), and cultured in an anaerobic environment at 37°C for 48 hours. After 48 hours, a single colony with typical morphology was scraped for The second passage, cultured under t...
Embodiment 2
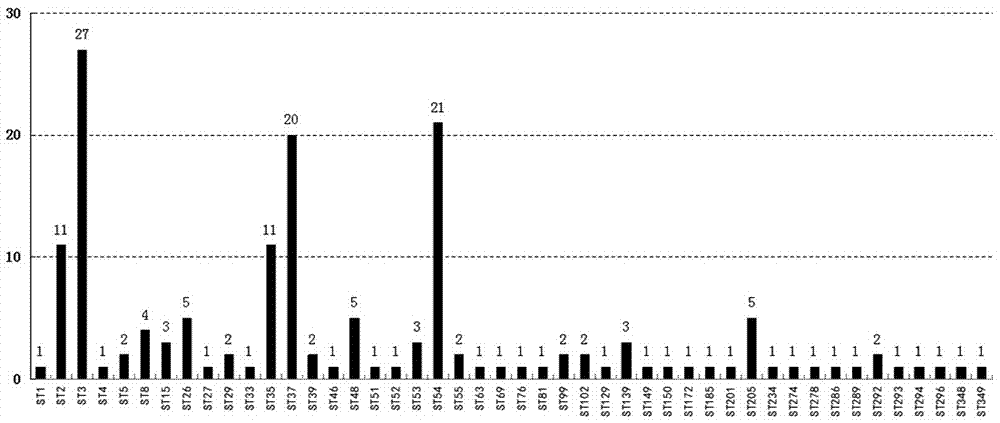
[0043] Samples: Randomly select 3 strains of Clostridium difficile ST35, 4 strains of Clostridium difficile ST2 and 3 strains of ST3 difficile, 1 C. difficile ST8, 4 C. difficile ST54, 1 C. difficile ST139, 1 C. difficile ST81 and 22 C. difficile ST37, all from HBCMCC .
[0044] Reagents used: with embodiment 1.
[0045] Instruments used: VITEK MS automatic rapid microbial mass spectrometry detection system, BioMérieux, France.
[0046] Each sample was tested as follows.
[0047] MALDI-TOF sample preparation: the method is the same as in Example 1.
[0048] data collection:
[0049] VITEK MS: Same as Example 1.
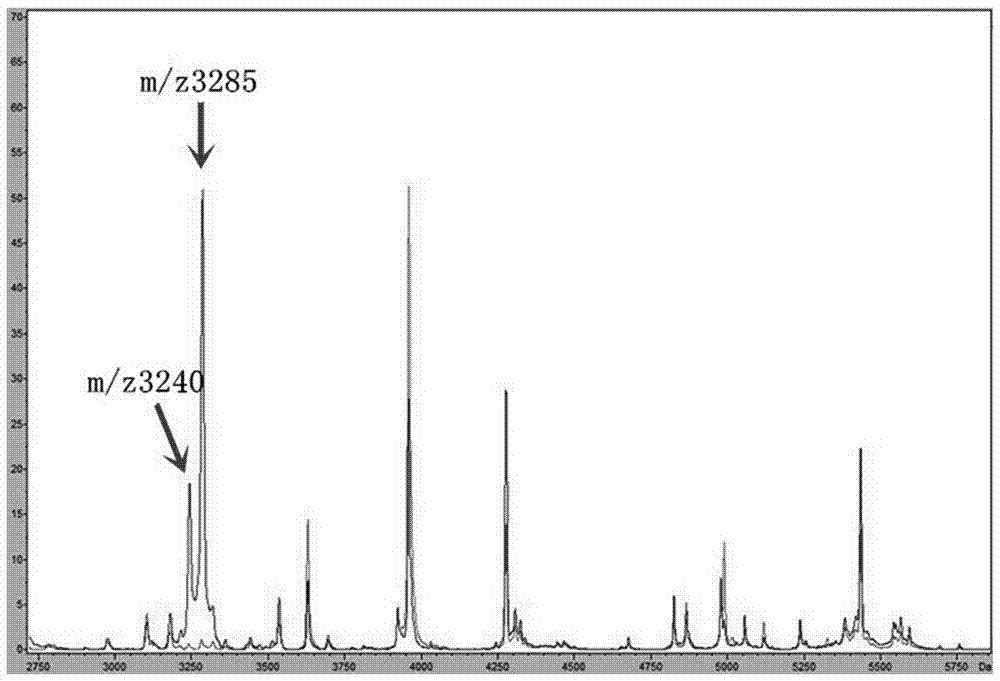
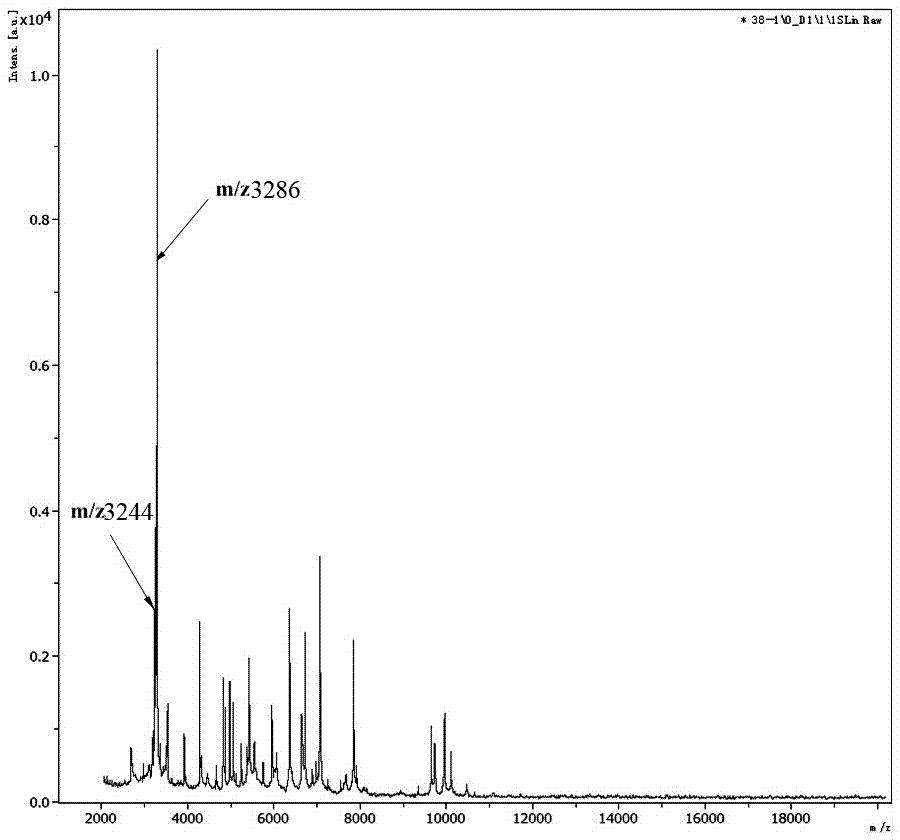
[0050] Data analysis: Detect whether there are protein peaks at m / z3235-3245 and m / z3280-3290 in the mass spectrum, or export a TXT file containing the peak list, and observe whether there are protein peaks at m / z3235-3245 and m / z3280-3290.
[0051] The identification results are shown in Table 1. The test results of 39 strains from different sources in the tabl...
Embodiment 3
[0057] Samples: 2 strains of Clostridium difficile ST37 (numbered 38 and 90 respectively) and 1 Clostridium difficile ST81 (numbered 81) were randomly selected from the strains of Clostridium difficile collected in different years (2010, 2012, 2014). ), 1 strain of Clostridium difficile ST3 (No. 3), 1 strain of Clostridium difficile ST102 (No. 45) and 1 strain of Clostridium difficile ST348 (No. 86), a total of 6 strains, all of which were from HBCMCC. See Table 2 for the strain number, type and year of collection.
[0058] Table 2 strain information
[0059]
[0060] The 6 strains of Clostridium difficile were inoculated and passaged twice at different times by three experimenters respectively, and the culture conditions were anaerobic culture at 37°C for 48 hours (anaerobic culture system, Guangzhou Youde Company). Randomly select three colonies in the medium of the second passage of each strain, and collect a total of 54 Clostridium difficile colonies as identification ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com