Dual-spacer sequence recognition and cleavage CRISPR-Cas9 vector construction, and application thereof in verrucosispora
A vector and sequence technology, applied in the field of genome editing, can solve the problems of long time, high cost, high off-target probability of CRISPR/Cas9 system, and achieve the effect of increasing stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0076] Example 1. Construction of a double spacer sequence recognition and cutting CRISPR-Cas9 vector and its application in Verrucospora
[0077] 1. Construction of double spacer sequence recognition cutting CRISPR-Cas9 vector
[0078] 1. Transform the carrier pHelp for auxiliary cloning spacer
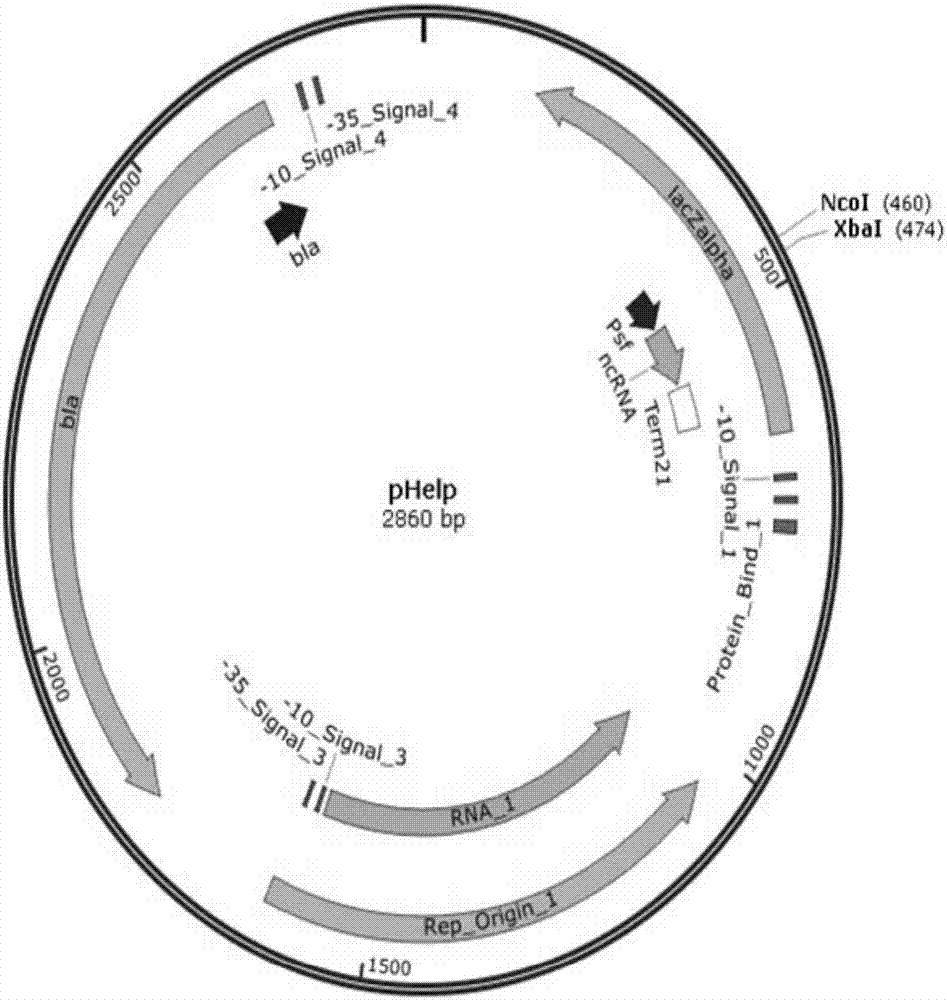
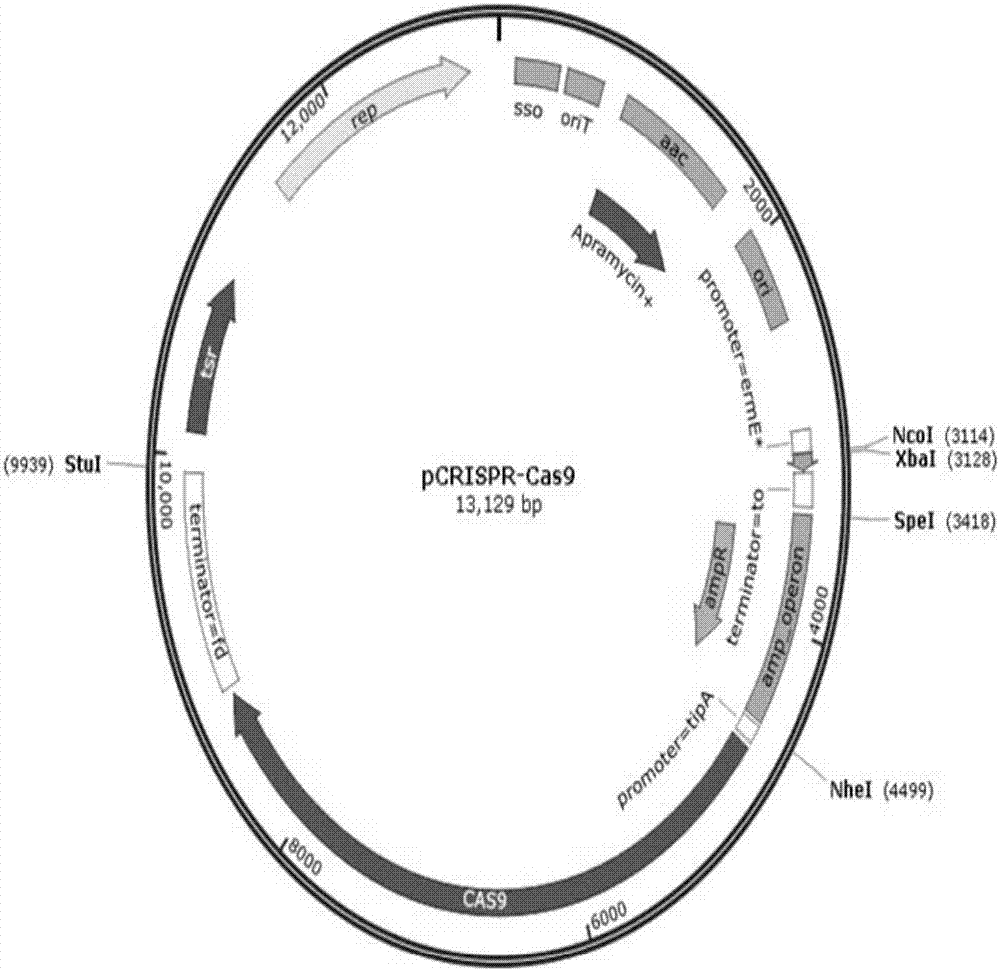
[0079] Transformation of the vector pHelp, which assists cloning and transcribing sgRNA, is carried out by a method including the following steps: Mutation assisting cloning expresses a spacer vector, adding a promoter psf and a terminator Term21 to it, and the promoter psf and terminator Term21 contain an element for transcribing sgRNA and enzyme digestion The recognition sites NcoI and XbaI can be inserted into the spacer sequence by means of sticky-end enzyme-cut ligation to obtain the transformed vector pHelp that assists in cloning the spacer (referred to as the transformed pHelp vector). The plasmid map is attached figure 1 , as can be seen from the figure: the modified pHelp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com