Method for degrading 1,2,3-TCP (trichloropropane) by engineered strain bacillus subtilis
A technology of Bacillus subtilis and engineering bacteria, applied in the biological field, can solve the problems of genetic instability, poor TCP tolerance, and low degradation efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Example 1 Obtaining Enzyme Genes and Relevant Elements Required for Construction of Integrated Vectors
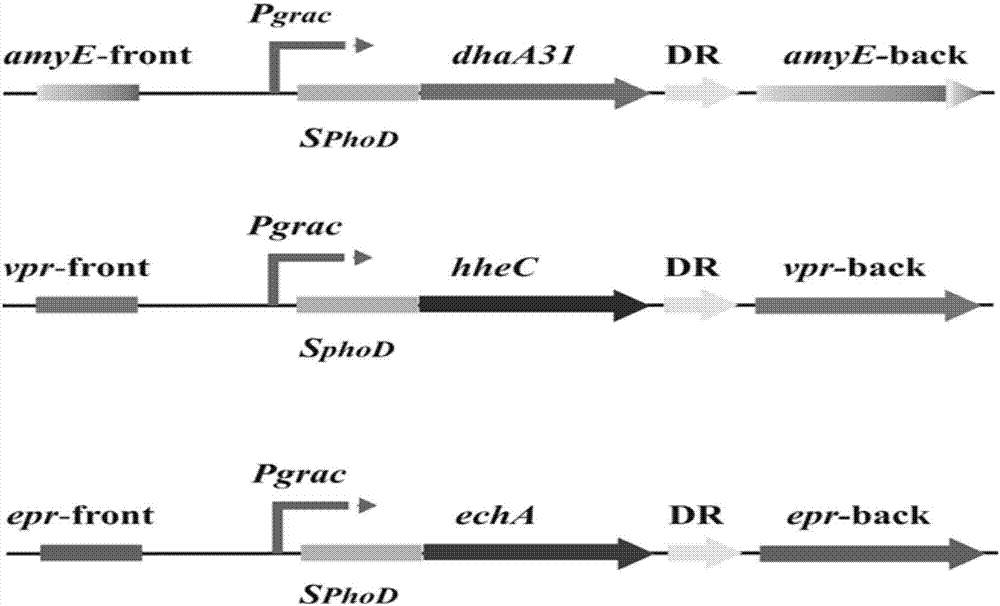
[0063] 1. According to the codon preference optimization of Bacillus subtilis, the haloalkane dehalogenase mutant gene was designed ( dhaA 31, the nucleic acid sequence is shown in SEQ ID NO.1), halohydrin dehalogenase gene ( wxya , the nucleotide sequence is shown in SEQ ID NO.3) and the epoxide hydrolase gene ( echA , the nucleic acid sequence is shown in SEQ ID NO.5).
[0064] The amino acid sequence of haloalkanedehalogenase (DhaA31) is shown in SEQ ID NO.2, the amino acid sequence of halohydrindehalogenase (HheC) is shown in SEQ ID NO.4, and the epoxide hydrolase ( epoxide hydrolase, EchA) amino acid sequence shown in SEQ ID NO.6.
[0065] 2. Insert the above-mentioned synthetic target gene into the plasmid pMD18-T to construct the recombinant plasmid pMD18T- dhaA 31. pMD18T- wxya and pMD18T- echA .
[0066] 3. PCR amplification to obtain the promoter...
Embodiment 2
[0080] Example 2 Construction of Bacillus subtilis integration vector
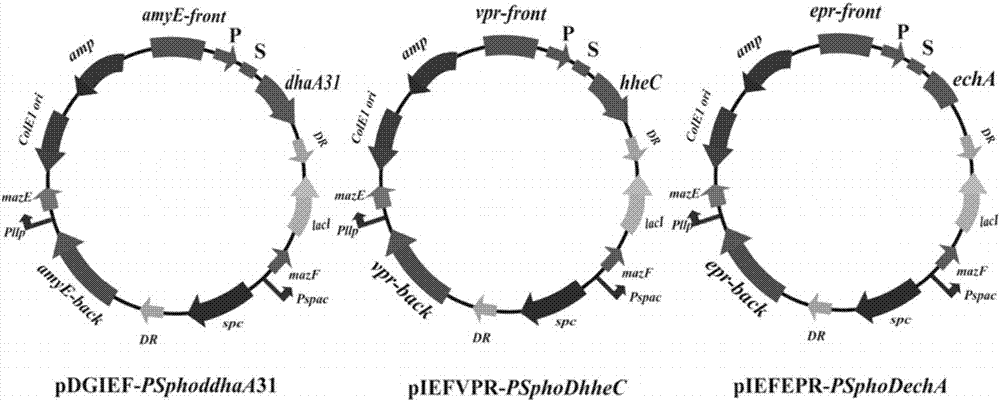
[0081] 1. Construction of recombinant plasmid pDGIEF- PSphoddhaA 31
[0082] (1) Download the gene sequence encoding plasmid pDGIEF (serial number: DQ358863.1) from the GenBank Data Base database, and synthesize the plasmid pDGIEF at Genaray Company (Shanghai Generay Biotech Co., Ltd), which is the host-wide plasmid pDGIEF.
[0083] (2) Using the recombinant plasmid pMD18T- dhaA 31 as template, apply primers dhaA 31-fw / dhaA 31 -rv Perform PCR amplification to obtain foreign genes dhaA 31; the amplified product was inserted into the plasmid pDGIEF after double digestion Sal I / not I site, construct the recombinant plasmid pDGIEF- dhaA 31.
[0084] The PCR program was as follows: 98°C, 10 min; 98°C, 10 s; 55°C, 10 s; 72°C, 15 s, for 30 cycles; 72°C, 3 min.
[0085] Primer dhaA31-fw (sequence shown in SEQ ID NO.14):
[0086] 5'-ATCCGC GTC GAC ATGTCAGAAATTGGCACAGGCTTTCCGTTTG-3' (the underlined...
Embodiment 3
[0148] Example 3 Construction of Bacillus subtilis engineering bacteria
[0149] 1. Transform the Bacillus subtilis integration vector constructed above into Escherichia coli competent cells, and extract the recombinant plasmid (such as figure 2 and 3 shown):
[0150] (1) Recombinant plasmid pDGIEF- PSphoddhaA 31 Transform Escherichia coli competent cells, extract the recombinant plasmid, and transform the recombinant plasmid pDGIEF- PSphoddhaA 31 After linearization, transform the competent cells of Bacillus subtilis, the recombinant plasmid and the host genome undergo double crossover homologous recombination, and integrate the target gene into the genome amye location. Select positive clones from the LB plate containing spectinomycin, inoculate them into 10 ml LB medium, culture at 37 °C, 200 rpm for 24 h, dilute the culture, and spread it on a solid LB plate containing 0.1 mM IPTG, Filter out IPTG s and spc r A single colony containing the exogenous gene dhaA 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com