Method of determining tumor marker based on transcriptome data
A tumor marker and data determination technology, applied in the field of bioinformatics, can solve the problems of difficulty in finding molecular markers for accurately predicting disease prognosis and complex regulatory systems from omics data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
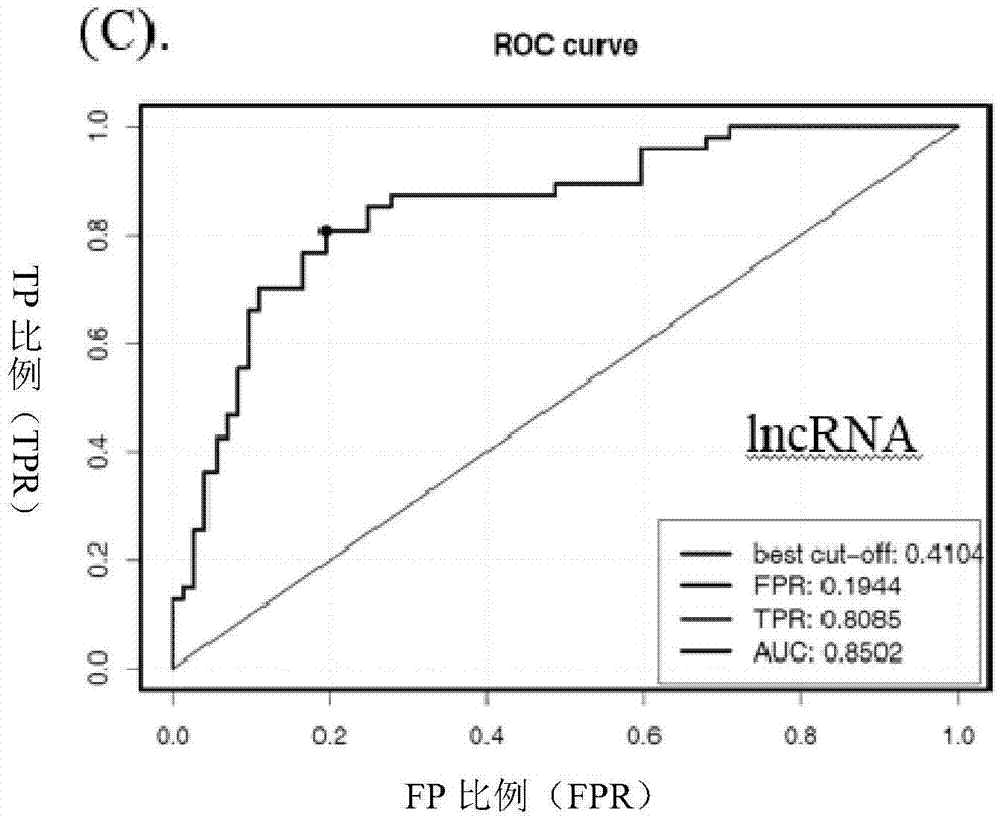
[0056] 1. Data description
[0057] mRNA, lncRNA and miRNA microarray expression data (Agilent RNA expression microarray) of normal tissue and tumor tissue of 119 ESCC samples.
[0058] 2. Data pre-processing
[0059] For miRNAs, the expression data of 208 miRNAs in 119 samples were selected after removing missing values; for lncRNAs, the probes annotated to specific data sets (UCSC, ENCODE, Cabili, etc.) were screened, followed by log transformation to screen for active expression and in For molecules differentially expressed in tumor and normal tissues, the expression data of 149 lncRNAs in 119 samples were finally obtained; for mRNA, the screening method was similar to lncRNA processing, and finally the expression data of 175 mRNAs in 119 samples were obtained. RNA expression data were normalized and used for subsequent analyses.
[0060] 3. Using group-lasso-based logistic for feature screening of miRNA, lncRNA and mRNA
[0061] 3.1 Sample grouping: 119 ESCC samples wer...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com