Fluorescence analysis method for determining concentration of Hg<2+>
A fluorescence analysis and concentration technology, which is applied in the field of fluorescence analysis to determine Hg2+ concentration, can solve the problems of complicated labeling process, insensitive fluorescence signal, unstable structure, etc., and achieves the effect of low cost, simple and effective operation, and enhanced fluorescence intensity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1 A method for measuring Hg 2+ Concentration Fluorescence Analysis Method
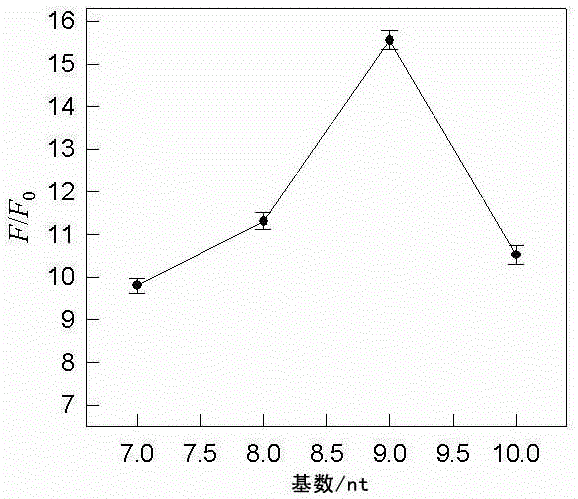
[0027] Select two non-labeled single-stranded DNA molecules A and B with different lengths and rich in T and G bases at the same time, and prepare solution A and solution B with a concentration of 100 μmol / L, respectively. 90°C water bath for 5 min, cooled to room temperature, mixed at a volume ratio of 1:1 to obtain a mixed solution, and set aside; wherein, the nucleotide sequence of the non-labeled single-stranded DNA molecule A is 5'-TTTTTTTGTGGGT-3', As shown in SEQ ID NO: 1; the nucleotide sequence of the non-labeled single-stranded DNA molecule B is 5'-GGGTGGGTGGTTATTTA-3', as shown in SEQ ID NO: 2; non-labeled single-stranded DNA molecules A and B The match length of is 7 T bases.
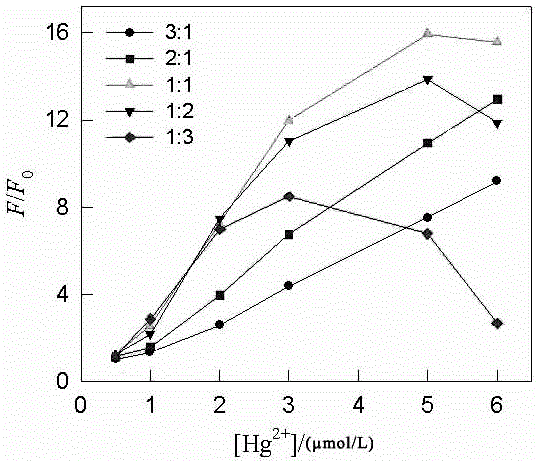
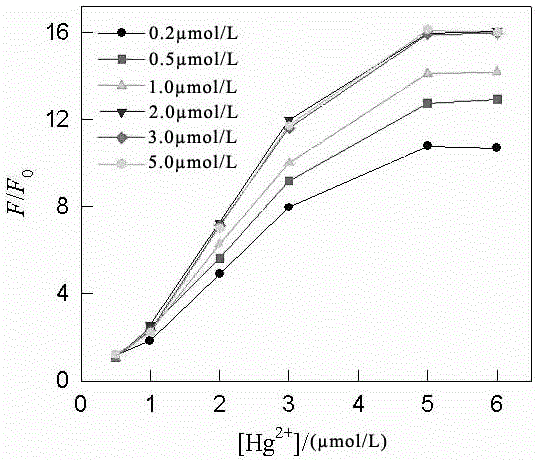
[0028] Mix 20 μL of the mixture with 10 μL of 200 μmol / L thioflavin solution to obtain mixture I; mix mixture I with 970 μL of 10 mmol / L Tris-HNO 3 The buffer solutions were mixed to obtain the mixe...
Embodiment 2
[0029] Embodiment 2 A method for measuring Hg 2+ Concentration Fluorescence Analysis Method
[0030]Select two non-labeled single-stranded DNA molecules A and B with different lengths and rich in T and G bases at the same time, and prepare solution A and solution B with a concentration of 100 μmol / L, respectively. Water bath at 90°C for 5 min, cooled to room temperature, and mixed at a volume ratio of 1:1 to obtain a mixed solution I for later use; wherein, the nucleotide sequence of the non-labeled single-stranded DNA molecule A is 5'-TTTTTTTTGTGGGT-3' , as shown in SEQ ID NO: 3; the nucleotide sequence of the non-labeled single-stranded DNA molecule B is 5'-GGGTGGGTGGTTATTTAT-3', as shown in SEQ ID NO: 4; the non-labeled single-stranded DNA molecule A and The matching length of B is 8 T bases.
[0031] Mix 20 μL of the mixture with 10 μL of 200 μmol / L thioflavin solution to obtain mixture I; mix mixture I with 970 μL of 10 mmol / L Tris-HNO 3 The buffer solutions were mixed...
Embodiment 3
[0032] Embodiment 3 A method for measuring Hg 2+ Concentration Fluorescence Analysis Method
[0033] Select two non-labeled single-stranded DNA molecules A and B with different lengths and rich in T and G bases at the same time, and prepare solution A and solution B with a concentration of 100 μmol / L, respectively. 90°C water bath for 5 minutes, cooled to room temperature, and mixed at a volume ratio of 1:1 to obtain a mixed solution I, which is set aside; wherein, the nucleotide sequence of the unlabeled single-stranded DNA molecule A is 5'-TTTTTTTTGTGGGT-3', As shown in SEQ ID NO: 5; the nucleotide sequence of the unlabeled single-stranded DNA molecule B is 5'-GGGTGGGTGGTTATTTATT-3', as shown in SEQ ID NO: 6; unlabeled single-stranded DNA molecules A and B The match length of is 9 T bases.
[0034] Mix 20 μL of the mixture with 10 μL of 200 μmol / L thioflavin solution to obtain mixture I; mix mixture I with 970 μL of 10 mmol / L Tris-HNO 3 The buffer solutions were mixed to ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com