Processing method and system for mitochondrial high-throughput sequencing data
A technology for sequencing data and processing methods, applied in the field of biological information, can solve the problem of inaccurate annotation of mitochondrial genomics, and achieve the effect of convenient clinical application and scientific research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
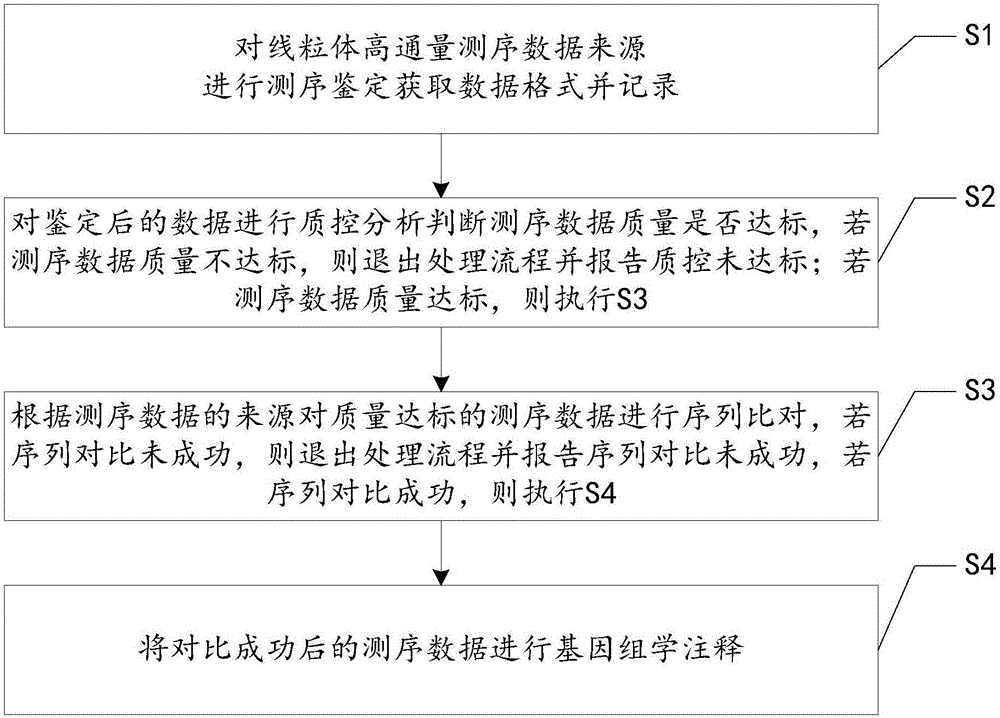
[0034] Such as figure 1 As shown, this embodiment proposes a method for processing mitochondrial high-throughput sequencing data, the method comprising:
[0035] S1. Sequence and identify the source of mitochondrial high-throughput sequencing data to obtain the data format and record it; if the sequencing data is in fastq format, it is Illnumina data; if it is in bam format, it is Ion Torrent data;
[0036] S2. Perform quality control analysis on the identified data to determine whether the quality of the sequencing data is up to standard. If the quality of the sequencing data is not up to standard, exit the processing flow and report that the quality control is not up to standard; if the quality of the sequencing data is up to standard, execute S3;
[0037] The quality control analysis of the data can be realized by using FastQC software. The input file required by this software is a question in the fastq format. Therefore, if the source of the data after sequencing is Illnum...
Embodiment 2
[0043] Such as Image 6 As shown, this embodiment proposes a mitochondrial high-throughput sequencing data processing system, the system comprising:
[0044] The data source judgment module is used to sequence and identify the source of mitochondrial high-throughput sequencing data to obtain and record the data format; if the sequencing data is in fastq format, it is Illnumina data; if it is in bam format, it is IonTorrent data;
[0045] The original data quality inspection module is used to perform quality control analysis on the identified data to determine whether the quality of the sequencing data is up to standard. If the quality of the sequencing data is not up to standard, it will exit the processing process and report that the quality control is not up to standard; if the quality of the sequencing data is up to standard, it will start Sequence comparison module;
[0046] The quality control analysis of the data can be realized by using FastQC software. The input file ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com