CRISPR (Clustered regularly interspaced short palindromic repeats)/Cas12 one-step nucleic acid detection method and 2019-nCoV detection kit
A technology for detection kits and detection methods, which is applied in the fields of microorganism-based methods, biochemical equipment and methods, and microorganism determination/inspection, etc., which can solve the problem that the results cannot be interpreted visually, the difficulty of popularizing basic inspection institutions, and the increase of complicated operations. problems, such as low cost, fast response, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
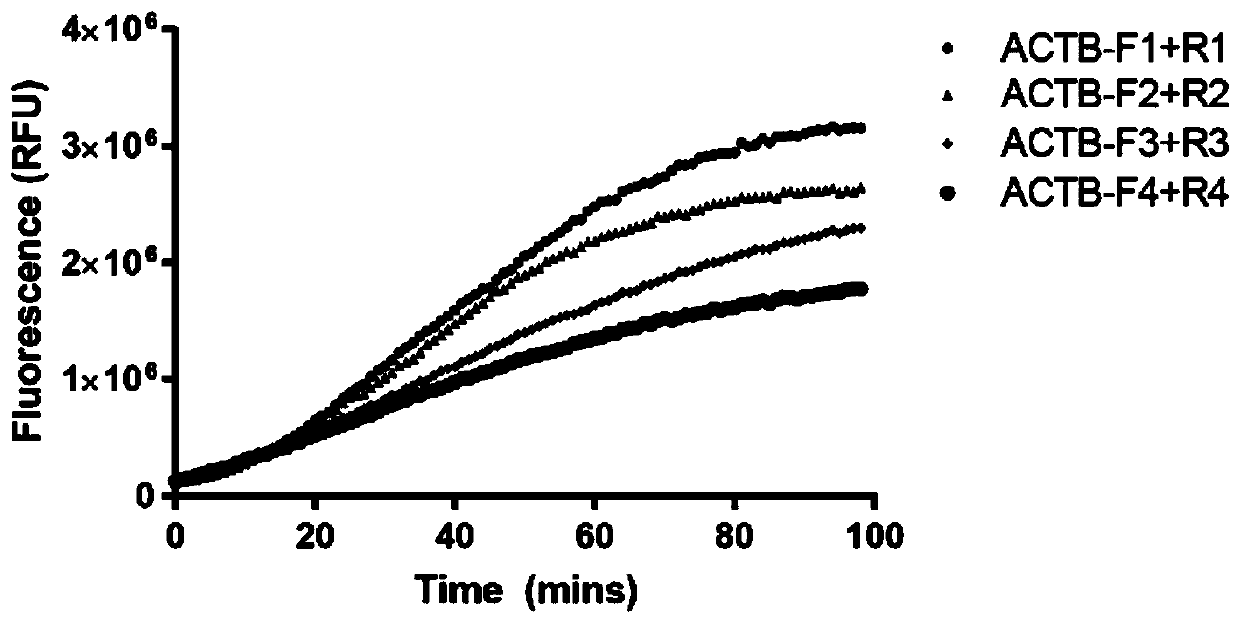
[0060] A CRISPR / Cas12 one-step nucleic acid detection method, which is a novel general-purpose CRISPR / Cas12-based one-step nucleic acid detection method, using crRNA, Cas protein, primers, buffer systems and single-stranded DNA reporter molecules, RNase inhibition components such as agents;
[0061] The following takes the detection of human β-actin (ACTB) as an example to describe the operation steps of the method:
[0062] 1) Search for the 5'-TTTN-3' sequence in the human β-actin region, and design a forward primer within 0-200bp near the 5' region, and design a reverse primer within 25-200bp near the 3' region . Synthesize and chemically modify the first three and last three phosphodiester bonds of the primers. The primers that can be used in this embodiment are shown in Table 1 below.
[0063] Table 1
[0064] Oligo name Sequence(5'-3') ACTB-F1 TGTGGATCAGCAAGCAGGAGTATGACGAGTCC (SEQ ID NO: 1) ACTB-F2 TGGATCAGCAAGCAGGAGTATGACGAGTCCGG (SEQ ID NO...
Embodiment 2
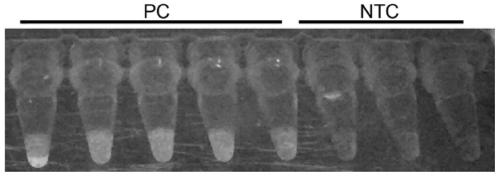
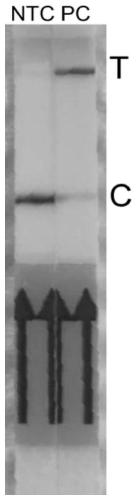
[0098] A new coronavirus detection kit, including crRNA, Cas protein, primers, buffer system and single-stranded DNA reporter; the comparison of the optimal primer combination of the above new coronavirus detection kit is determined as follows:
[0099] 1) Search for the 5'-TTTN-3' sequence in the conserved ORF1ab and N gene regions of the new coronavirus, and design forward primers within 0-200 bp of the near 5' region, and 25-200 bp in the near 3' region Design reverse primers. Synthesize and chemically modify the first three and last three phosphodiester bonds of the primers. The primers that can be used in this embodiment are shown in Table 11 below.
[0100] Table 11
[0101] Oligo name Sequence(5′-3′) ORF1ab-F1 TTGCCTGGCACGATATTACGCACAACTAATGGT (SEQ ID NO: 11) ORF1ab-F2 ATGGTGACTTTTTGCATTTCTTACCTAGAGTTT (SEQ ID NO: 12) ORF1ab-R AAGTCAGTGTACTCTATAAGTTTTGATGGTGTGT (SEQ ID NO: 13) N-F1 CCAGGCAGCAGTAGGGGAACTTCTCCTGCTAGAAT (SEQ ID NO:...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com