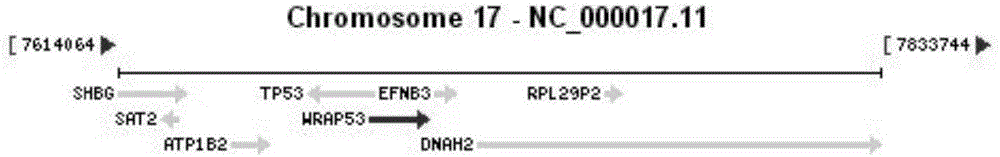
Method and primers for detecting dyskeratosis congenita (DC)-related gene WRAP 53
A gene, wrap53-exon2-f technology, applied in the field of gene mutation primers, which can solve the problems of few gene detection and small number of cases, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0115] A primer for detecting the polymorphic mutation site of the WRAP53 gene, the design of the primer is an amplification primer designed for the whole exon of WRAP53, including:
[0116] The primers for amplifying the whole exon sequence of WRAP53 gene, its base sequence is:
[0117] WRAP53-Exon1A-F: TGTAAAACGACGGCCAGTGGGAACGGGAAACCTTCTAA
[0118] WRAP53-Exon1A-R: AACAGCTATGACCATGGACAGCAGTCCGGAGCTAAC
[0119] WRAP53-Exon1B-F: TGTAAAACGACGGCCAGTCTAATCTCCGCTGTGCTTCC
[0120] WRAP53-Exon1B-R: AACAGCTATGACCATGTCTTCTGCAGGAAGGCTTGT
[0121] WRAP53-Exon1C-F: TGTAAAACGACGGCCAGTGGGACCCAGTTTCTCTCTCC
[0122] WRAP53-Exon1C-R: AACAGCTATGACCATGCTGGAGAAGTGGGTCTCAGG
[0123] WRAP53-Exon2-F: TGTAAAACGACGGCCAGTGTGGAGTCTGGGGAGATGAA
[0124] WRAP53-Exon2-R: AACAGCTATGACCATGGGGCATCCCTCTCCTAGAAA
[0125] WRAP53-Exon3-F: TGTAAAACGACGGCCAGTCAGCCCTAGCCCTACACTTG
[0126] WRAP53-Exon3-R: AACAGCTATGACCATGTGCTGCCACAAGAAATTCAC
[0127] WRAP53-Exon4-F: TGTAAAACGACGGCCAGTTCTGAGCTCACCCTTGAACA
...
Embodiment 2
[0153] Operation process of blood / cell / tissue genomic DNA extraction kit (Tiangen Biology):
[0154] (1) Genomic DNA extraction from whole blood
[0155] 1) Add 20μl QIAGEN Protease (or proteinase K) to the bottom of a 1.5ml centrifuge tube. .
[0156] 2) Add 200 μl of plasma to the centrifuge tube.
[0157] 3) Add 200 μl Buffer AL and shake for 15 seconds. (Note: Do not add QIAGEN Protease or proteinase K directly to Buffer AL. If the sample size is large, increase QIAGEN Protease and Buffer AL proportionally.)
[0158] 4) Water bath at 56°C for 10 minutes, and then briefly centrifuge to remove the liquid on the inner edge of the centrifuge tube cover.
[0159] 5) Add 200 μl of ethanol (96%-100%), shake for 15 s, and briefly centrifuge to remove the liquid along the inner edge of the centrifuge tube cover.
[0160] 6) Carefully add the mixture obtained above (including the precipitate) into the QIAamp micro spin column (do not wet the edge of the spin column). Put the sp...
Embodiment 3
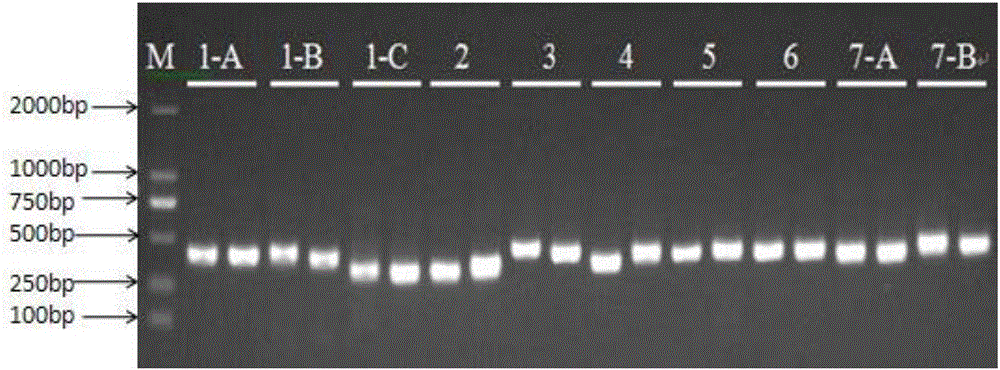
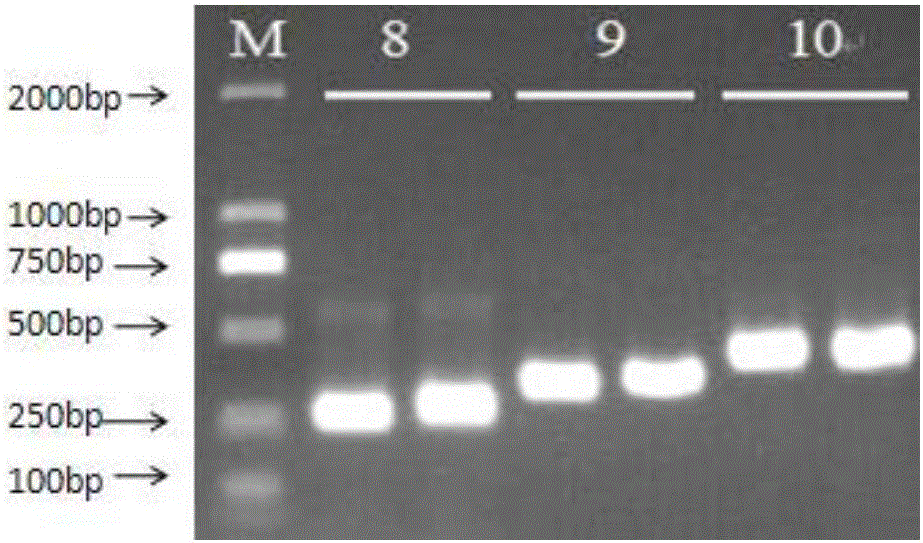
[0217] In order to verify the feasibility of the primers and the entire detection system, 13 cases of clinical samples were taken (two repetitions were established for each pair of primers, and each group was numbered 1-13 by primers) according to the reagents and methods of Examples 1 and 2 to extract genomes and prepare reagents , amplification and sequencing. Take sample 4 and add primers for amplification, and the electrophoresis results are as follows: figure 2 and image 3 As shown, it shows that the primers of the present invention can effectively amplify blood samples, and the band is single. The sequencing results are shown in the table below.
[0218]
[0219]
[0220] Figure 4 It shows the front and back sequencing screenshots of the WRAP53 exon 2 wild type of sample 4, indicating that the peaks of the sequencing results are clear and there are no interferences such as miscellaneous peaks and heavy peaks that affect the results.
[0221] It can be seen f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com