Rapid screening method for Lactobacillus helveticus rich in ace-inhibiting active peptides, combined sequence and construction method for realizing the method
A construction method and screening method technology, which is applied in the field of joint sequence, rapid screening of Lactobacillus helveticus rich in ACE-inhibiting active peptides, can solve the problems of Lactobacillus helveticus lacking ACE active peptides, and achieve low screening cost, simple operation, High throughput effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Activation, cultivation and identification of embodiment 1 bacterial strain
[0065] The samples of Lactobacillus helveticus (Lactobacillus helveticus) that have been analyzed for the characteristics of ACE-inhibiting active peptides in the lactic acid bacteria strain resource bank of Inner Mongolia Agricultural University were used as the analysis objects, and a total of 12 strains were collected. The strain information is as follows:
[0066] Table 2 Isolation sources and ACE inhibitory peptide characteristics of Lactobacillus helveticus strains
[0067]
[0068] Do the following for each sample:
[0069] Activation of bacterial strains: Put the freeze-dried bacterial powder stored in the ampoule tube into skim milk for activation and rejuvenation (37°C, 24h), microscopically examine the bacterial liquid smear, and the microscopic examination results are Gram-positive pure cultures Continue to subculture to the third generation.
[0070] Use liquid nitrogen free...
Embodiment 2
[0071] The construction of embodiment 2 joint sequence
[0072] 2.1 Design primers
[0073] Using Lactobacillus helveticus H9 (genbank number CP002427) as a template, the key genes of pepX, pepN and pepO involved in the proteolysis system were selected as targets. Primer Premier 5.0 software was used to design specific amplification primers, and each primer was artificially synthesized as shown in Table 1 above.
[0074] 2.2
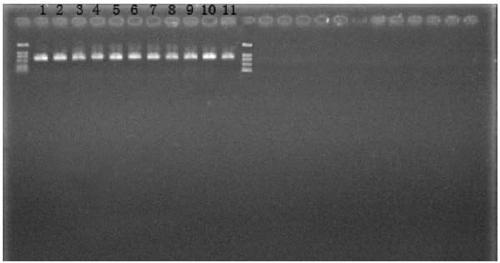
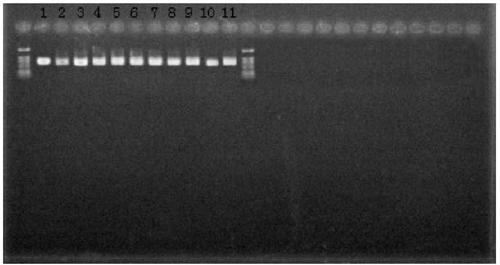
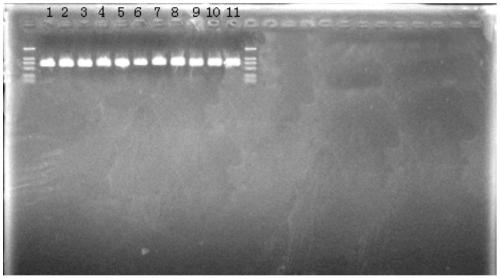
[0075] The primers in Table 1 were used to amplify the template DNA by PCR, respectively, to obtain three gene fragments of each strain, and the template DNA came from the 12 isolates in Table 2.
[0076] Amplification system: template DNA 50ng, 10×PCR buffer 5μL, dNTPs (2.5mmol / μL) 4μL, upstream and downstream primers 10μM each 1.5μL, Taq DNA polymerase (5U / μL) 0.5μL, ddH 2 O to make up to 50 μL.
[0077] Amplification conditions:
[0078] Pre-denaturation at 95°C for 5min; denaturation at 95°C for 1min, annealing at 50°C for 45s, extension at 72°C...
Embodiment 3
[0083] Example 3 Construction of developmental tree
[0084] Using the disclosed Lactobacillus helveticus H9 genomic DNA as a template, amplified in the same steps as in Example 2, the resulting gene fragments were concatenated in the order of pepX-pepN-pepO to obtain the combined gene sequence of SEQ No.1, Used as a standard for screening methods.
[0085] The sequence of each strain of Example 2 was concatenated in the order of pepX-pepN-pepO to obtain the combined gene sequences of 12 strains to be screened respectively, and the nucleic acid sequences thereof are shown in SEQ No.2-13.
[0086] Adopt MEGA6.0 software package to analyze joint sequence, through Clustal W alignment, the joint gene sequence of 12 bacterial strains that embodiment 2 isolates obtains and the joint sequence of SEQ No.1 carries out pairwise comparison, calculates the distance between the sequences, Construct a distance matrix to reflect the genetic relationship between sequences.
[0087] Then con...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com