Reagent system and kit for detection of colorectal cancer spindle assembly checkpoint and application
A kit and spindle technology, applied in the field of colorectal cancer spindle assembly checkpoint target gene detection, can solve the problems of low accuracy, limited application, time-consuming and laborious, and achieve low misdiagnosis rate, high sensitivity and specificity. strong effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] The steps to obtain a DNA preservation solution for a stool sample include:
[0061] 1. Collect fecal samples from subjects exposed for less than 24 hours, and pick 170-180 mg of samples on ice with a sterile razor blade.
[0062] 2. Add 3.0ml of feces lysate to the sample, centrifuge after fully lysing, take the supernatant and add 250mg of bovine serum albumin, centrifuge to obtain the supernatant, add 30μl of proteinase K to the supernatant and place it at 60°C The digestion reaction was carried out for 2 hours to obtain the digestive juice.
[0063] The composition of the stool lysate is: 30mmol / L Tris-Cl, 3mmol / L EDTA, 0.6mmol / L NaCl and 0.1% (W / V) sodium lauryl sulfate.
[0064] 3. Cool the digested solution obtained above to room temperature, add 300 μl protein precipitation solution to precipitate protein, mix well, remove protein impurities by centrifugation, and obtain supernatant, which is DNA extract.
[0065] The protein precipitation solution is prepared...
Embodiment 2
[0069] The steps to obtain a DNA preservation solution for a stool sample include:
[0070] 1. Collect fecal samples exposed for less than 24 hours, and pick 200-220 mg of samples on ice with a sterile razor blade.
[0071] 2. Add 0.5ml of feces lysate to the sample, centrifuge after fully lysing, take the supernatant and add 250mg of bovine serum albumin, centrifuge to obtain the supernatant, add 30μl of proteinase K to the supernatant and place it at 60°C The digestion reaction was carried out for 2 hours to obtain the digestive juice.
[0072] The composition of the stool lysate is: 100mmol / L Tris-Cl, 10mmol / L EDTA, 5mmol / L NaCl and 1.0% (W / V) sodium lauryl sulfate.
[0073] 3. Cool the digested solution obtained above to room temperature, add 300 μl protein precipitation solution to precipitate protein, mix well, remove protein impurities by centrifugation, and obtain supernatant, which is DNA extract.
[0074] The protein precipitation solution is prepared from 2mol / L p...
Embodiment 3
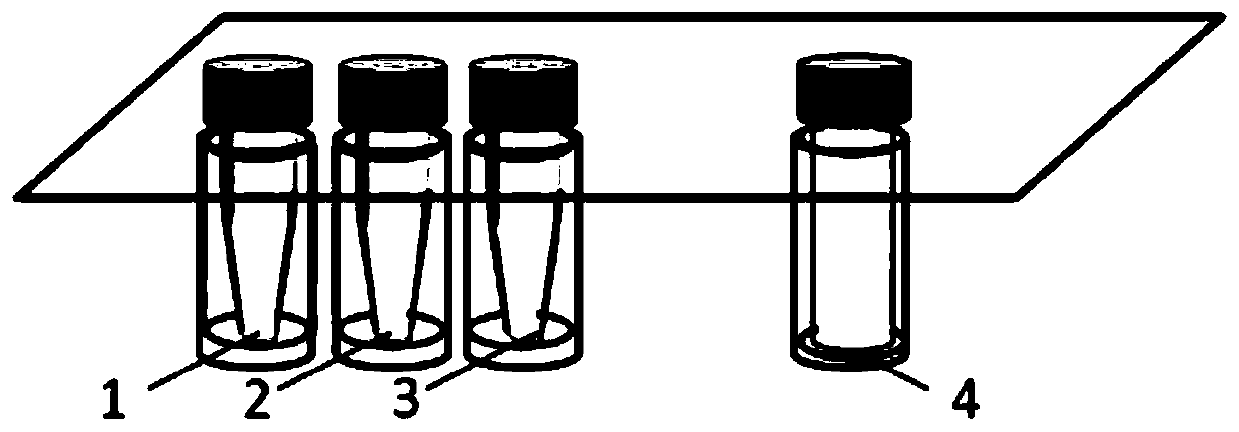
[0078] Prepared kit as attached Figure 1-3 As shown, except for the packing box, it mainly includes two parts, the upper layer and the lower layer. The upper part is used to obtain the DNA preservation solution of the stool sample, wherein, 1 is equipped with a stool lysate, 2 is equipped with a silica gel membrane binding solution, 3 is equipped with a silica gel membrane washing solution, and 4 is equipped with a protein precipitation solution.
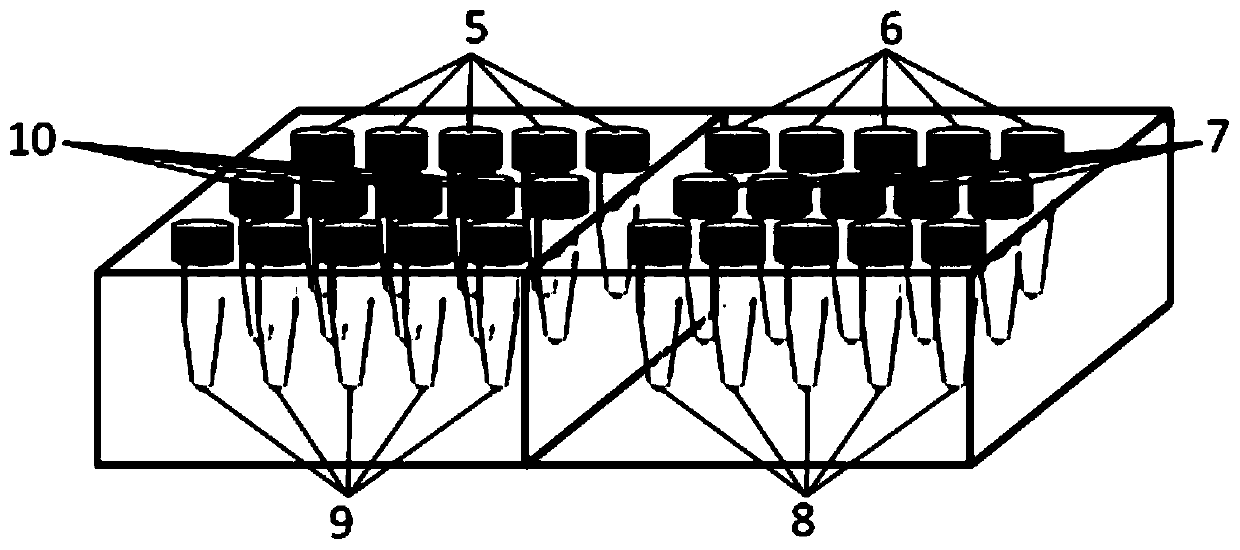
[0079] The lower part contains reagents for each gene detection, among which, 5 is a reagent for BubR1 detection, 6 is a reagent for Mad1 detection, 7 is a reagent for Mad2 detection, 8 is a reagent for PlK1 detection, 9 is a reagent for AURKB detection, 10 is a reagent for Cdc20 detection Use reagents. Each group of 5 PE tubes contains DNA amplification primers corresponding to six genes, PCR reaction components, TaqDNA polymerase, PCR product purification components and DNA probe sequencing reaction components.
[0080] After u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com