Group of NZ2114 histidine mutants, and preparation method thereof
An amino acid, a series of technologies, applied in the field of protein engineering and genetic engineering, can solve the problems such as histidine, mutation, etc. that have not yet been seen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Embodiment 1 NZ2114 histidine mutant molecular design and gene synthesis
[0040] Based on the presence of two histidines at the 16th and 18th positions in the NZ2114 sequence, arginine and lysine were used to mutate them. A total of 5 sequences (SEQ ID No. 1-SEQ ID No. 5) were designed for single-site mutants and double-site mutants.
[0041] The mutant gene sequences (SEQ ID No.6-SEQ ID No.10) were designed according to the codon preference of Pichia pastoris. In order to ensure the integrity of the sequence during expression, an XhoI restriction site and a Kex2 restriction site were added to the 5' end of the mutant gene sequence, and a TAA, TAG terminator sequence and XbaI restriction site were added to the 3' end. The above sequence was completed by Shanghai Sangon Bioengineering Co., Ltd.
Embodiment 2
[0042] Example 2 Construction of Pichia pastoris inducible expression vector pPICH1~pPICH5
[0043] The synthetic gene fragment and vector pPICZaA were double-digested with restriction endonucleases XhoI and XbaI respectively, and the pPICZaA vector fragment and mutant gene fragment were recovered and ligated to obtain vectors pPICH1-pPICH5;
[0044] Detailed construction process of vectors pPICH1~pPICH5: use restriction endonucleases XhoI and XbaI to carry out double enzyme digestion on the synthesized gene fragment and pPICZaA respectively. The enzyme digestion system and conditions are as follows:
[0045]
[0046]
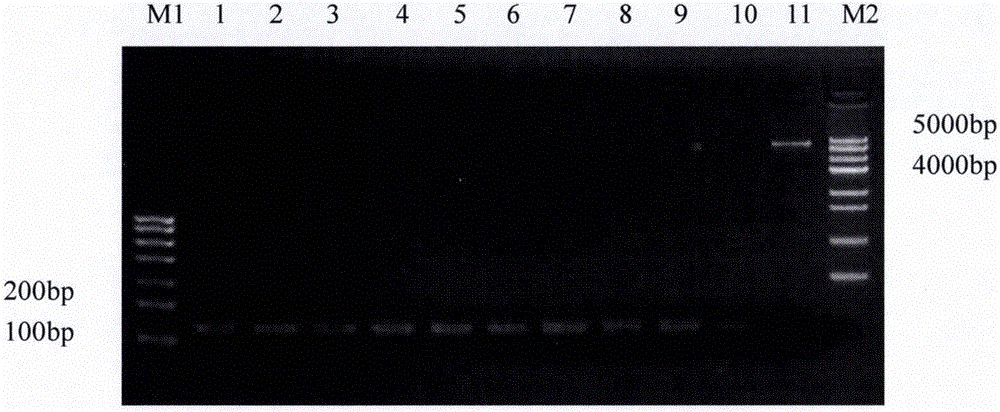
[0047] After adding the sample to the above enzyme digestion system, react at 37°C for 3 hours, and detect by 2% agarose gel electrophoresis, electrophoresis conditions: 120V, 30min. After electrophoresis, use a scalpel to cut the electrophoretic bands corresponding to the carrier fragment and the gene fragment under the ultraviolet light, and use the Tia...
Embodiment 3
[0063] Example 3 Construction of recombinant yeast strain containing pPICH1~pPICH5
[0064] 3.1 Linearization of recombinant vector pPICH1~pPICH5
[0065] Use PmeI to digest the constitutive recombinant expression vectors pPICH1~pPICH5. The enzyme digestion system and reaction conditions are as follows:
[0066]
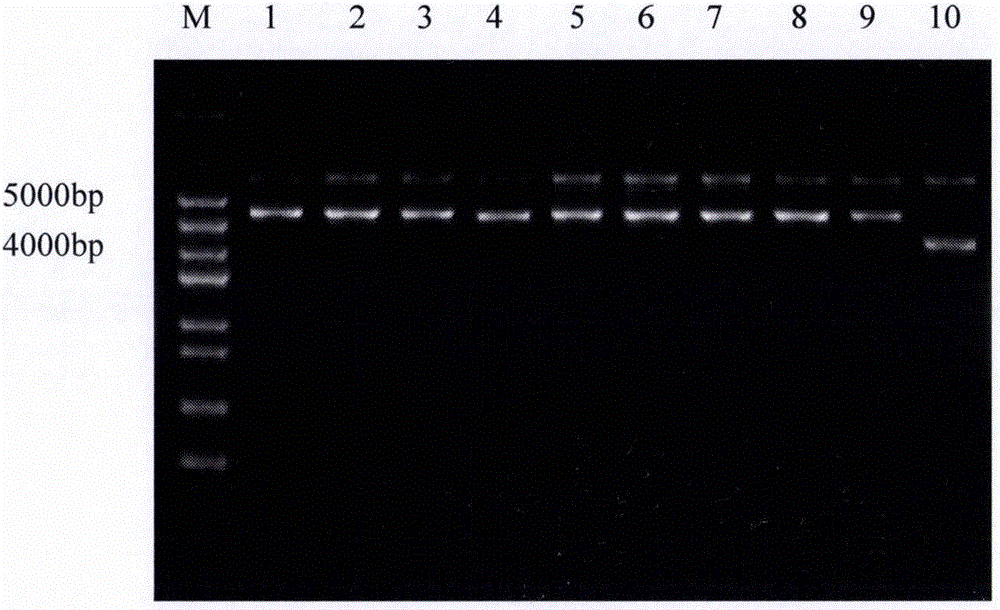
[0067] After adding the sample to the above enzyme digestion system, react at 37°C for 3 hours, and detect by 2% agarose gel electrophoresis, electrophoresis conditions: 120V, 30min. After the electrophoresis is completed, it is detected that the recombinant expression vector is correctly linearized, and then the linearized recombinant expression vector is recovered using a DNA recovery kit.
[0068] 3.2 Pichia pastoris electrotransformation and identification of linearized vector
[0069] 1) Pick a single colony of X-33 on the YPD plate, inoculate it into 10mL YPD liquid medium, culture at 30°C, 250rpm overnight;
[0070] 2) Inoculate 50μL overnight culture so...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| The inside diameter of | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com