Construction method suitable for 16S rDNA high-throughput sequencing library
A sequencing library and construction method technology, applied in the field of 16SrDNA high-throughput sequencing library construction, can solve problems such as insufficient coverage of amplified sequences, low library recovery efficiency, and cumbersome steps, so as to shorten the time for library construction and save libraries Effect of purification time and simplified operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
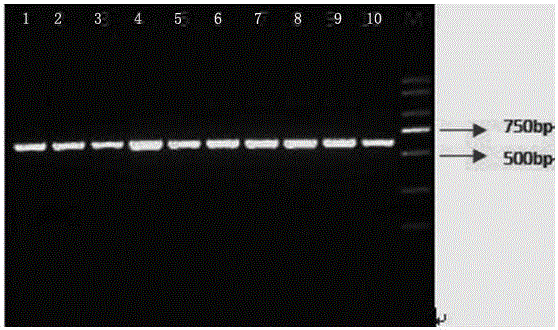
Image
Examples
Embodiment 1
[0025] (1) Collect 10 cases of human alveolar lavage fluid samples, about 2ml per sample, centrifuge at room temperature, 14000rpm, 30min, discard the supernatant after centrifugation, and save the precipitate. Using the QIAamp® UCP Pathogen Mini kit from Qiagen, the genomic DNA was extracted according to the protocol.
[0026] (2) Use nanodrop 2000 to detect the DNA concentration and quality, and select DNA with a concentration greater than 10 ng / ul and OD260 / 280=1.8-2.0 for subsequent library construction operations, with a total of 10 DNA samples.
[0027] (3) The primers used in the PCR amplification of 10 samples were randomly combined with the aforementioned 8 upstream primers and 12 downstream primers, as shown in the following table:
[0028]
[0029] (4) Using Takara Premix Ex Taq TM Hot Start Version for PCR amplification.
[0030] The PCR reaction system is as follows:
[0031] Premix Ex Taq TM Hot Start Version 25μl
[0032] Forward primer (10μM) 1μl
[...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com