Molecular marker detecting ischemic cerebralvascular accident and application thereof
A technology of ischemic stroke and molecular markers, applied in the field of biomedicine, can solve the problem of lack of molecular markers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] The collection of embodiment 1 sample
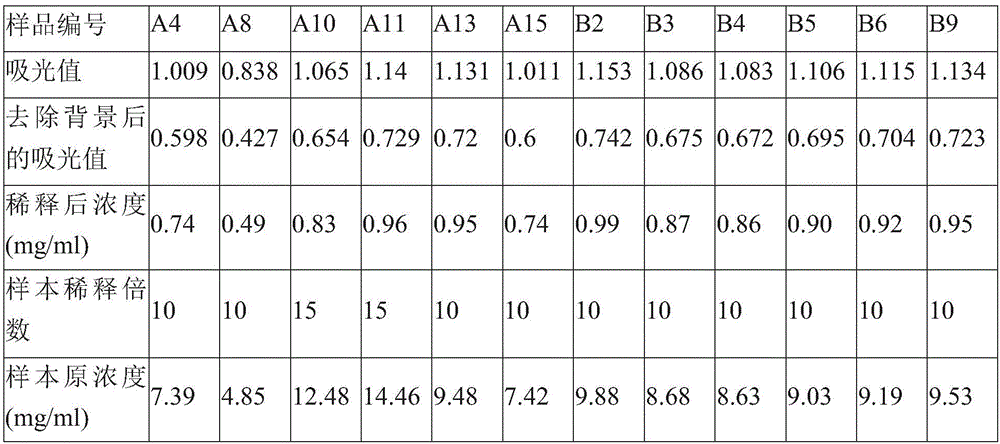
[0055] From June 2015 to December 2015, patients with ischemic stroke who were treated in PLA Rocket Army General Hospital were collected. A total of 6 cases were collected in the case group. A total of 6 healthy volunteers were collected, and the specific information is shown in Table 1. Obtain 5ml of cubital median venous blood from all research subjects, put it in a 4-degree refrigerator for 1 hour, centrifuge at 1500 rpm for 10 minutes, take the supernatant, centrifuge at 1500 rpm for 10 minutes, take the supernatant, and store in a -80-degree refrigerator.
[0056] Table 1 Sample information
[0057] factor
Embodiment 2
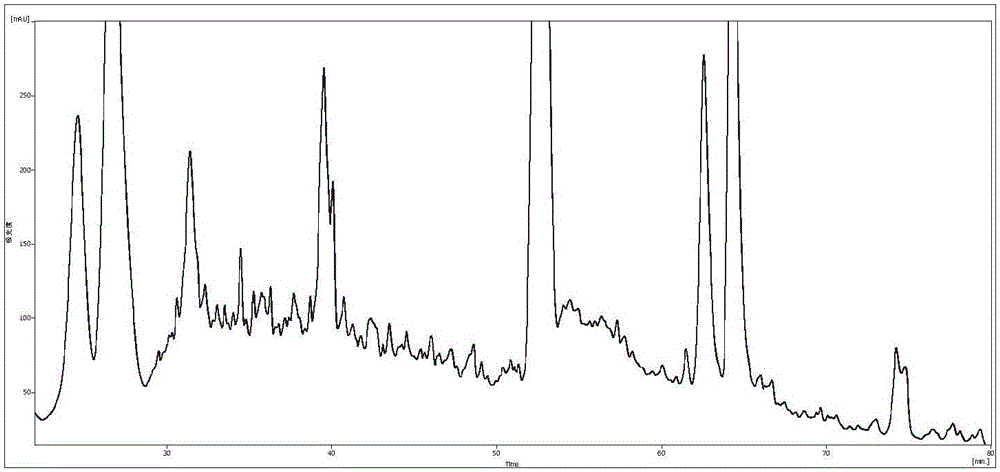
[0058] Example 2 iTRAQ experiment screening differential proteins
[0059] 1. Instruments and reagents used
[0060] Vortex oscillator (Haimen Qilinbeier Instrument Manufacturing Co., Ltd., model: QL-901)
[0061] Constant temperature incubator (Shanghai Pudong Rongfeng Scientific Instrument Co., Ltd., model: HH.S4)
[0062] Vacuum freeze dryer (Thermo, model: SPD2010-230 )
[0063] RIGOL L-3000 HPLC system (Beijing Puyuan Jingdian Technology Co., Ltd.), mobile phase A: 98% ddH2O, 2% acetonitrile (pH 10); mobile phase B: 98% acetonitrile, 2% ddH 2O (pH 10) high performance liquid chromatography: (ThermoScientic EASY-nLC 1000 System (Nano HPLC)), mobile phase A: 100% ultrapure water, 0.1% formic acid; mobile phase B: 100% acetonitrile, 0.1% formic acid mass spectrometry System (Thermo, model: Q-Exactive )
[0064] Blue Albumin
[0065] IgG Removal Kit (Sigma-Aldrich, Cat. No.: PROTBA-1KT )
[0066] DTT (Bio-Rad, catalog number: 161-0611, USA )
[0067] Iodoacetamide (B...
Embodiment 3
[0133] Example 3 Large sample verification of expression of differential protein PIGR
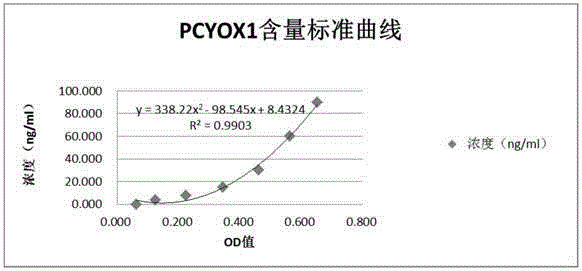
[0134] (1) Dilution of the standard substance Add 1mL of the standard substance diluent to each bottle of the standard substance, cover it and let it stand at room temperature for about 10 minutes. Prepare 7 EP tubes of diluted standard, add 150 μL of standard diluent to each EP tube, dilute into different gradients, and directly use the standard diluent (0pg / mL) as a blank well.
[0135] (2) Adding samples: set up blank wells (blank control wells do not add samples and enzyme-labeled reagents, and the rest of the steps are the same), standard wells, and sample wells to be tested. Accurately add 50 μl of the standard substance on the enzyme-labeled plate, add 40 μl of the sample diluent to the well of the sample to be tested, and then add 10 μl of the sample to be tested (the final dilution of the sample is 5 times). Adding the sample Add the sample to the bottom of the well of the microti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com