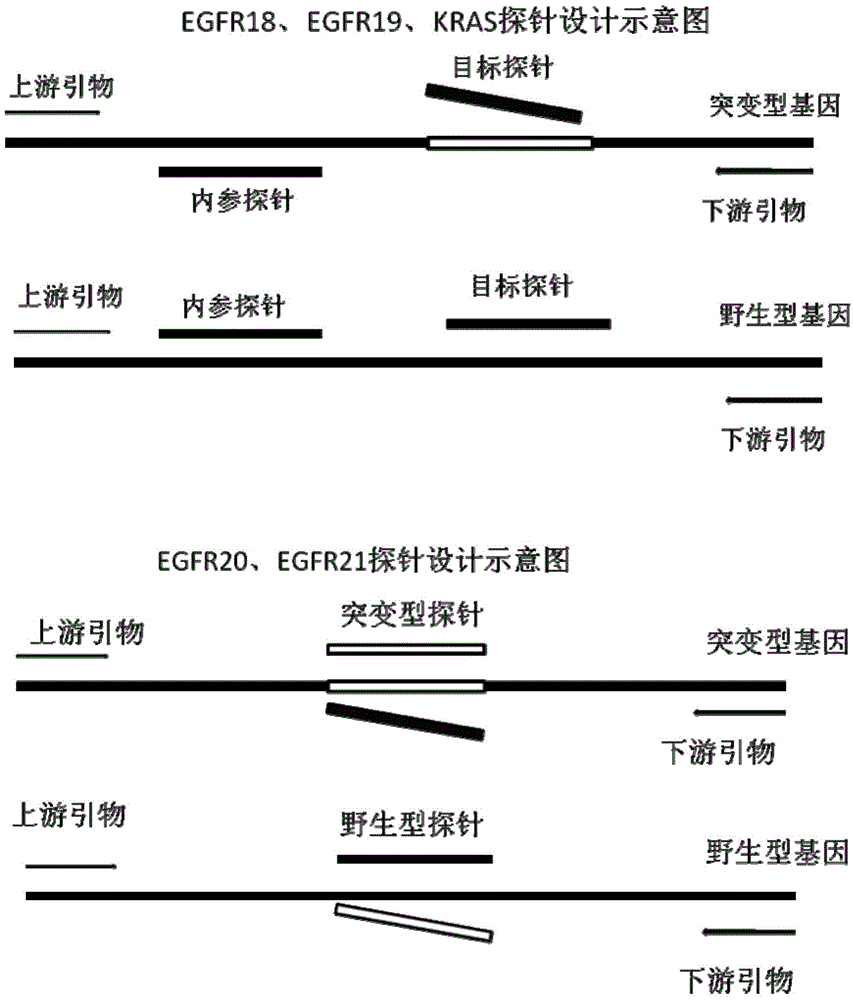
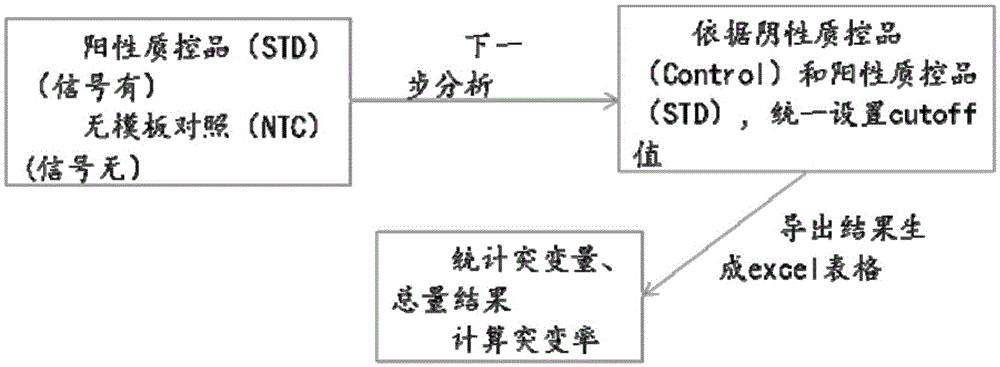
Primer, probe and kit for detecting EGFR and/or K-ras genetic mutation
A probe and gene technology, applied in the field of devices for detecting gene mutations based on a digital PCR platform, can solve the problems of complex sample types, limited detection content, and inability to completely block the amplification of wild-type DNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0136] Embodiment 1: Composition and method of use of the kit of the present invention
[0137] The composition of the kit of the present invention is shown in Table 4 above, and its usage method is also described in the summary of the invention, and the detailed steps are shown in the following examples.
Embodiment 2
[0138] Embodiment 2: Simulation sample detection
[0139] (1) Prepare the sample: mix the DNA containing the wild-type EGFR gene with the positive quality control in a certain ratio. The source of DNA can be serum, plasma, peripheral blood, oral mucosa, pleural effusion, body fluid or tissue, etc.
[0140] In this example, samples A, B, and C were obtained by mixing different amounts of the above-mentioned positive plasmids with the same amount of wild-type DNA. Before mixing, the positive plasmids and wild-type DNA were respectively quantified using a digital PCR instrument to obtain a theoretical mutation rate. .
[0141] (2) Melt PCR primers and corresponding TaqMan probes at room temperature.
[0142] (3) Prepare the PCR reaction solution according to the ratio of the kit instructions.
[0143] (4) Add 20 μL of the prepared digital PCR mixture to the 8-lane droplet making plate, and then add 60 μL of droplet making oil to the making plate to prepare PCR micro-reaction d...
Embodiment 3
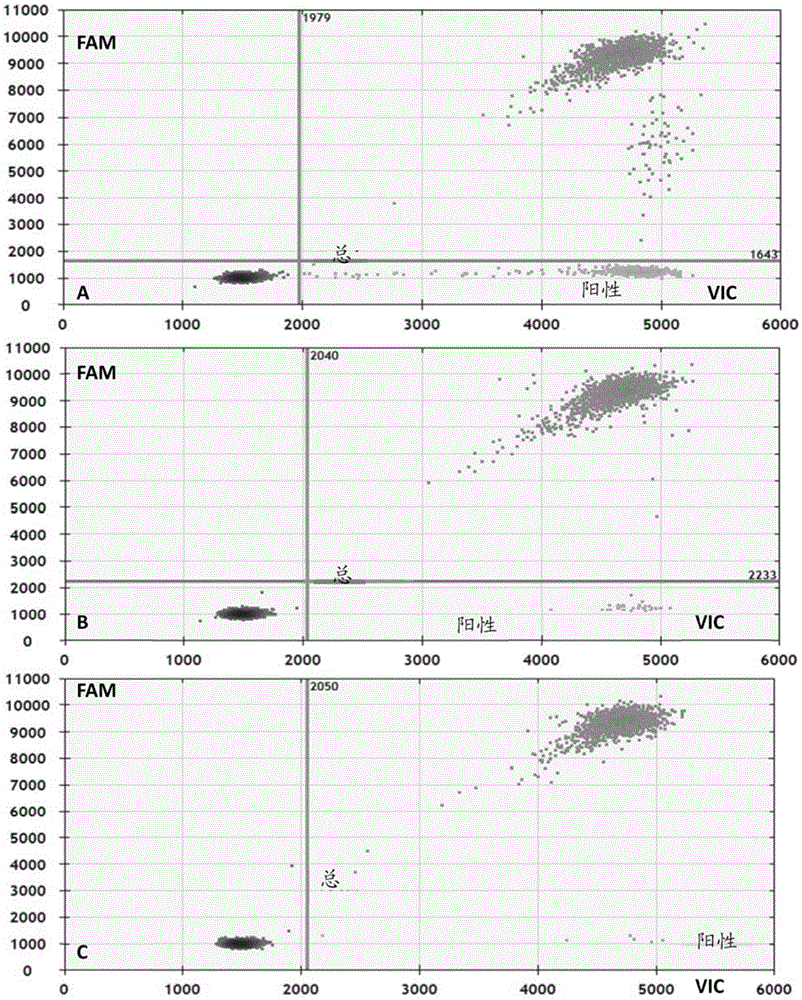
[0150] Embodiment 3: detection of paraffin-embedded standard substance
[0151] According to the usage method of the above-mentioned embodiment 1, the samples were taken as the standard product QuantitativeMultiplexFFPEReferenceStandard (purchased from Horizon Diagnostics Co., Ltd.). Test results such as Figure 4 shown. The analysis of the detection results is the same as in Example 1, the mutation rate=1-(Number wild type / Number total molecule)=1-(NumberFAM(+)VIC(+) / NumberVIC(+)), the specific results are derived, the wild-type signal The number and the total number of signals are as follows:
[0152]
[0153] The results of quantitative analysis show that the error is generally 0.3%-2.5%, not more than 5%. With the above method, it is possible to detect c.2235-c.2253 deletion mutations on exon 19 of EGFR, and various types of point mutations c.35, c.34, and c.38 on exon 2 of KRAS gene, and the detection limit reaches 2%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com