Novel alkali-resistant glycoside hydrolase family 10 xylanase produced from micobacterium sp. HY-17 strain
A technology of xylanase and bacterial bacteria, which is applied in the field of xylanase, can solve environmental problems such as harmful
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Production of xylanase (Xylanase) Bacillus (Micobacterium sp.) isolation of bacterial strains
[0086] The present inventors isolated microorganisms having enzymatic activity for hydrolyzing xylan from commensal microorganisms in the intestines of mole crickets.
[0087]Specifically, in order to selectively isolate microorganisms that hydrolyze xylan, M9mineralsaltsagarmedium (Difco) was used as a minimal medium, and a selective medium containing 5g / Lyeastextract (Difco) and 2g / Lazo-xylan (Megazyme) was used . In the selection medium, after diluting the microbial suspension in physiological saline by serial dilution method, inject 100 μl into each selection medium, and incubate at 25-35° C. for 18-36 hours. In each medium, a colony of microorganisms forming a transparent ring due to the decomposition of azo-xylan was selectively isolated. In order to identify the isolated microorganism, PCR on the 16S rDNA sequence was performed with a pair of 27F primer (SEQ ID NO: ...
Embodiment 2
[0090] Cloning of Xylanase (Xylanase)
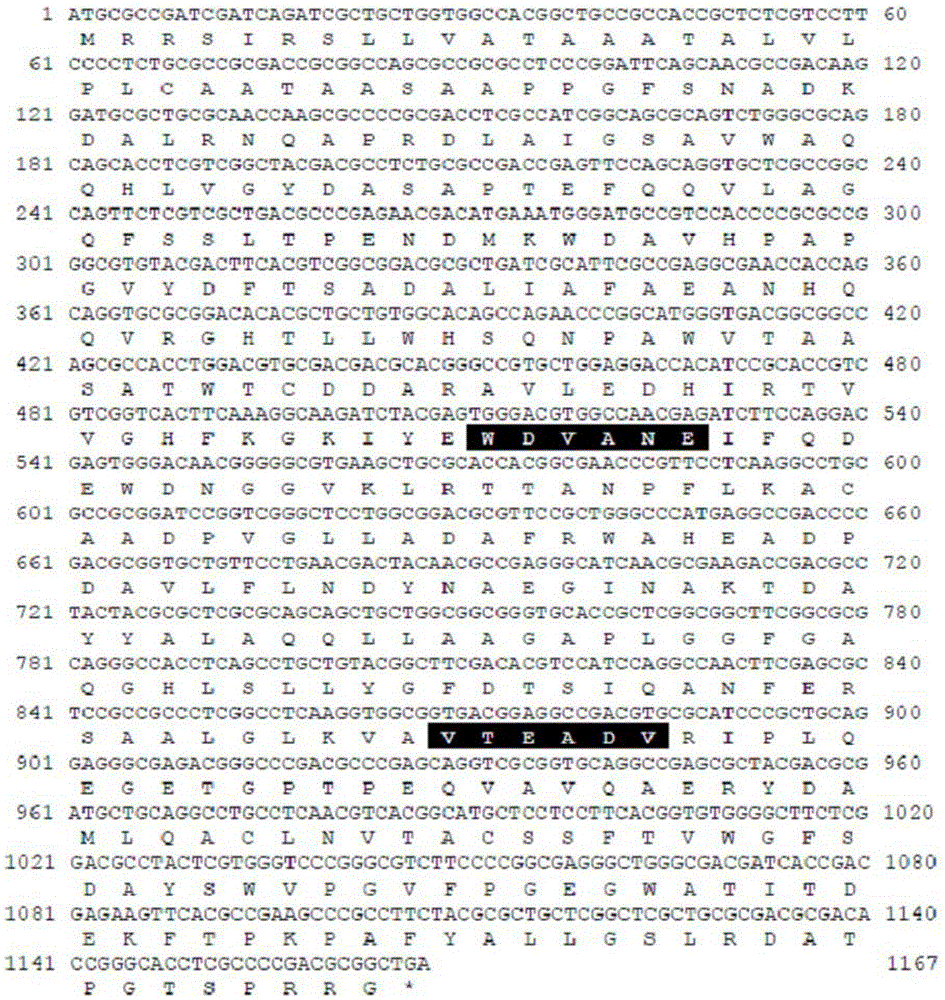
[0091] In the genomic DNA of the Bacillus (Micobacteriumsp.) HY-17 bacterial strain (preservation number: KCTC12338BP) that decomposes xylan, utilize the gene sequence based on the xylanase gene sequence of GH10 (glycosidehydrolasefamily10) series, amplify The polynucleotide sequence encoding the xylanase protein was amplified and cloned.
[0092] Specifically, after the genomic DNA was isolated from the strain, using the genomic DNA as a template, using 10× buffer (MgCl2), 2.5mMdNTPs, 5× buffer, FastStartTaq DNA polymerase (Roche) and MF-10 forward The primer (SEQ ID NO: 3) was paired with the MR-10 reverse primer (SEQ ID NO: 4), and PCR for the xylanase gene was performed. At this time, the PCR conditions were denaturation at 95°C for 5 minutes, denaturation at 95°C for 30 minutes, annealing at 50°C for 30 seconds, and stretching at 72°C for 40 seconds. A final stretch was performed for 7 minutes at 72°C. The PCR product of the 375...
Embodiment 3
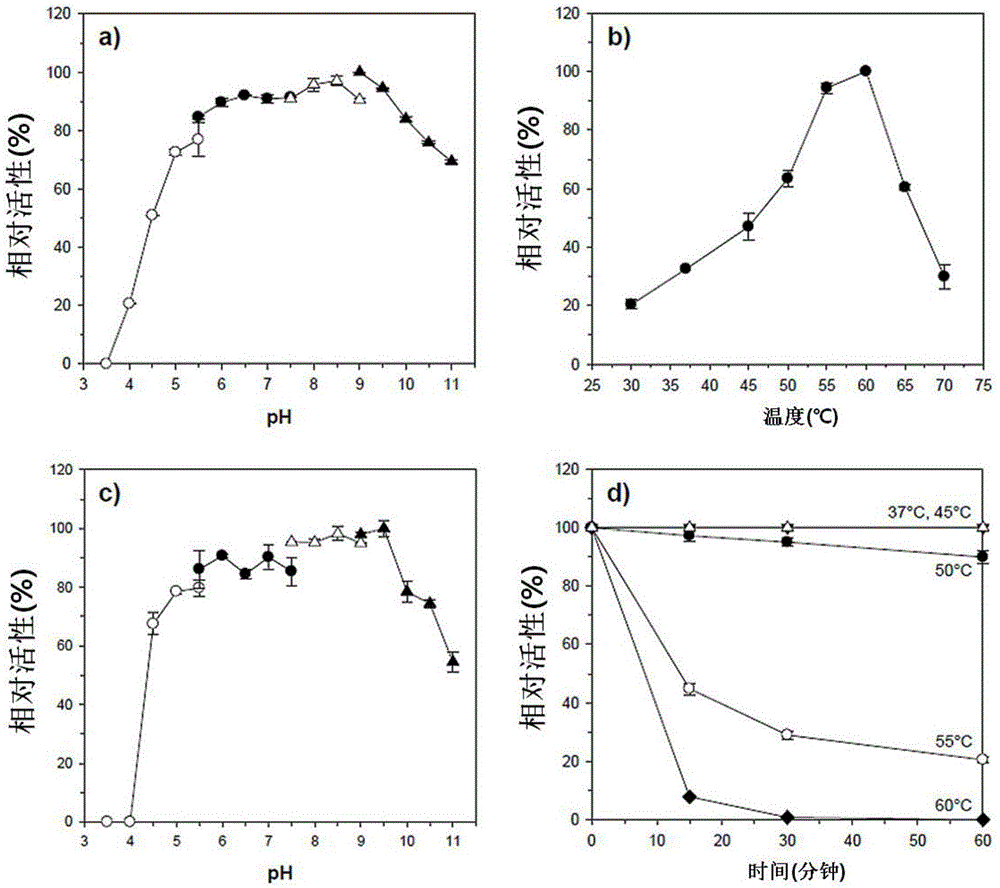
[0098] Refining of xylanase (Xylanase)
[0099] Recombinant XylH (rXylH) obtained by overexpressing the pET-XylH expression vector produced in in Escherichia coli was isolated.
[0100] Specifically, Escherichia coli transformed with each expression vector was inoculated into a liquid LB medium, and cultured with shaking at 37°C. When the OD600 value of each Escherichia coli culture solution reached 0.4 to 0.5, 1.0 mM IPTG was added, followed by shaking and culturing at 30° C. for 5 hours. The culture solution was centrifuged, and the result observed after the cells were pulverized by a sonic pulverizer, rXylH activated inclusion bodies with 20 mM sodium phosphate buffer solution (sodium phosphate buffer pH 7.4) containing 20 mM imidazole (imidazole) and 0.5 M sodium chloride. (inclusion bodies) soluble. The solubilized rXylH cell crush was refolded and purified using a HisTrapHP (GE Healthcare, Sweden) (5-ml) column, and then a liquid chromatography (LC) system (Amphamasi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com