Pair of TALENs for efficiently editing rice WAXY gene, and identification targeting site and application thereof
A technology for identifying components in rice, applied in the fields of biotechnology and genetic engineering, can solve problems such as low efficiency, limited gene knockout, and undetectable genetic modification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0201] Example 1 TALENs target sequence design
[0202] 1. Download the rice (Oryza sativa) WAXY gene from NCBI, and design 5 pairs of TALENs combinations. The WAXY gene sequence is shown in Appendix 1.
[0203] 2. TALENs recognition sites are designed according to the following general principles:
[0204] (1) The 0th base is T, that is, the base before the first in the recognition sequence is the 0th, which is T;
[0205] (2) The length of the spacer sequence (Spacer) between the two recognition sequences is between 15-30;
[0206] (3) The length of the recognition sequence is between 15-24.
[0207] 3. The designed TALENs target sequence recognition sites and spacer regions are as follows. The underline is the TALEN recognition binding region, and the middle is the spacer region.
[0208]
Embodiment 2
[0209] Example 2 TALENs construction
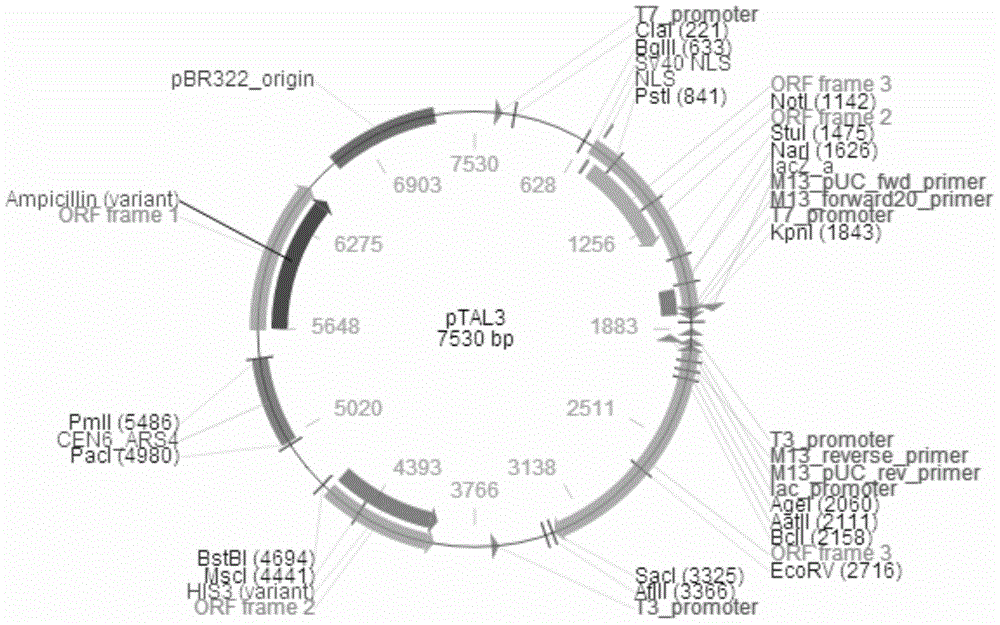
[0210] According to the classic GoldenGate method (Cermak, T. etal. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research, 2011, volume. 39, No. 12e82 doi: 10.1093 / nar / gkr218) for TALE assembly, NI, NG, HD, NN four modules were identified A , T, C, G four bases. The combined modules were constructed on the background plasmid pTAL3 vector (purchased from Addgene, USA), and the sequence was verified to be correct by sequencing. pTAL3 vector such as figure 1 shown.
Embodiment 3
[0211] Example 3 TALENs expression vector construction
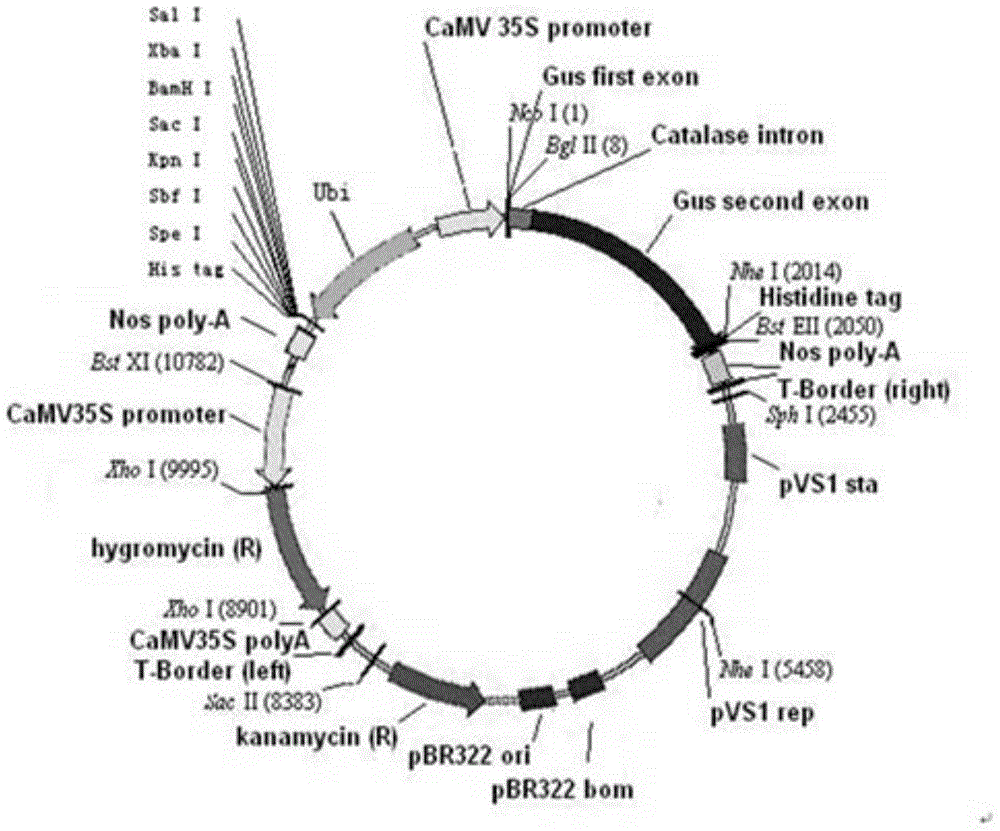
[0212] The TALENs coding region in the correctly sequenced pTAL3 vector was connected to the transformed plant expression vector pCMBIA1301 using KpnI and SpeI, BglII and BstEII respectively (pCMBIA1301 was purchased from the China Plasmid Vector Strain and Cell Line Gene Collection Center, and pCMBIA1301 was transformed as follows: ubi+ The MCS+nos expression cassette (sequence SEQ ID NO.: 30) was added to the pCMBIA1301 vector using HindIII and EcoRI restriction enzymes). Finally, the expression vector contains a selection gene—hygromycin gene, and a pair of TALENs are placed under the CaMV35S (cauliflower mosaic virus) promoter and Ubi (maize ubiquitin 1) promoter, such as image 3 shown. For plant expression vector plasmid information, see figure 2 .
[0213] The ubi+MCS+nos expression cassette sequence is shown in SEQ ID NO.:30.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com