SNP molecular markers used in chromosome 6 of pig for traceability and application thereof
A technology of molecular markers and chromosomes, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, DNA/RNA fragments, etc., can solve the problems that molecular markers cannot be independent of each other, individuals cannot be distinguished, and molecular marker linkage phenomena, etc., to achieve perfect detection Effects of coverage, improved accuracy, and expanded detection range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Example 1 Search for Molecular Markers
[0030] (1) Primer design
[0031] Using the DNA sequence of pig chromosome 6 (Genbank: FN673727) as a template, design primers to isolate the DNA fragment of pigs (using the DNA of a Duroc pig as a template for PCR amplification), the primers are as follows:
[0032] Forward primer: 5'-TCAGTCAGTGCCGTTTCA-3' (as shown in SEQ ID NO.2)
[0033] Reverse primer: 5'-GACAGGCAGCAATGTTAGG-3' (as shown in SEQ ID NO.3)
[0034] The total volume of the PCR reaction is 20μl, including about 100ng of porcine genomic DNA, containing 1×buffer (Promega Company), 1.5mmol / LMgCl 2 , the final concentration of dNTP (Shanghai Sangon Biological Company) was 150 μmol / L, the final concentration of primers was 0.2 μmol / L, and 2UTaq DNA polymerase (Promega Company).
[0035] PCR amplification: 94°C for 4min, (94°C for 30s, 60°C for 30s, 72°C for 40s) cycled 30 times, and finally 72°C for 10min.
[0036] PCR reaction products were detected by 1% agarose...
Embodiment 2
[0047] The distribution situation of embodiment 2 alleles
[0048] (1) Design of test groups
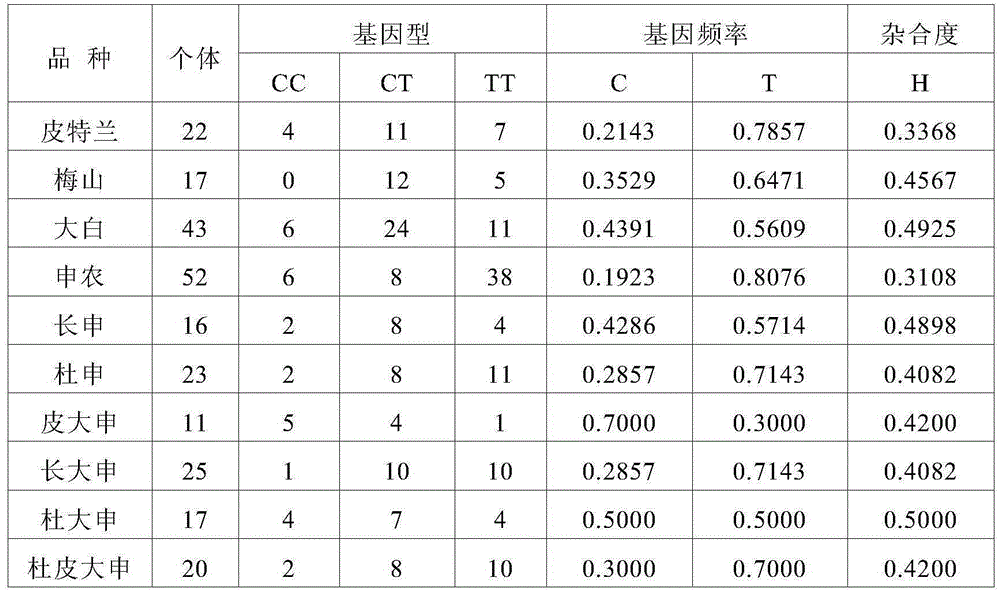
[0049] Experimental group: collected Pietrain (21 heads), Meishan (17 heads), Dabai (43 heads), Shannon (52 heads), Changshen (16 heads), Dushen (23 heads), Pi Dashen (11 heads) ), Dachangshen (25 heads), Du Dashen (17 heads) and Du Pi Dashen (20 heads) individual ear tissues, and DNA was extracted, a total of 246 DNA samples.
[0050] The purpose of the test population is to detect the distribution of SNP markers in different breeds.
[0051] (2) Genetic testing
[0052] Forward primer: 5'-TCAGTCAGTGCCGTTTCA-3' (as shown in SEQ ID NO.2);
[0053] Reverse primer: 5'-GACAGGCAGCAATGTTAGG-3' (as shown in SEQ ID NO.3).
[0054] Perform amplification (amplification conditions are the same as in Example 1), and use the same PCR-Eco72I-RFLP method to detect all individual genotypes of the test population.
[0055] (3) Statistical analysis
[0056] Record all individual genotypes of th...
Embodiment 3
[0060] Example 3 SNP molecular marker usability traceability test verification
[0061] The SNP molecular marker of the present invention has been preliminarily confirmed that it can be used in traceability markers. In order to ensure the usability in traceability markers, the present invention re-sampled in different locations, and existing 13 SNP molecular markers (as shown in Table 2 below) Shown) combined use (total 14 SNP molecular markers) to carry out usability traceability test verification, at the same time, use only the existing 13 SNP molecular markers to carry out traceability test verification as a comparison example.
[0062] Randomly collect 100 ear tissue samples and 100 muscle samples from individual pigs at Fuxing Slaughterhouse (every pig individual sample is collected at the same time as ear tissue samples and muscle samples), ear tissue samples are numbered E1-E100, and muscle samples are numbered M1-M100, respectively DNA from muscle samples and ear tissu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com