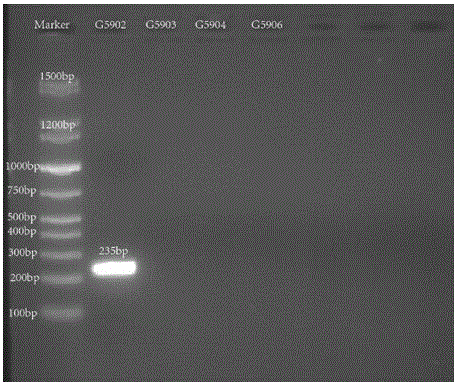
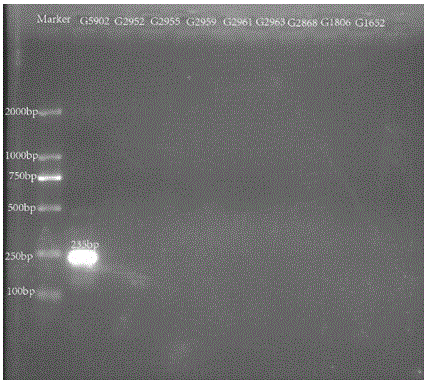
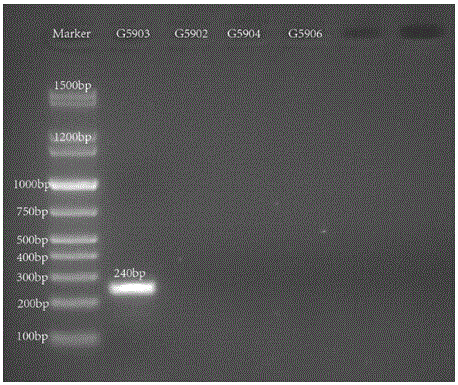
Nucleotide specific to hafinia alvei G5902, G5903, G5904 and G5906, and application of nucleotide
A Hafnia bacterium and nucleotide technology, which is applied in the determination/testing of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of low sensitivity, high missed detection rate, and long time-consuming serotyping methods, etc. problem, to achieve the effect of low detection cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 : Genome Extraction
[0038] Hafnia alvei was cultured in nutrient broth medium at 37°C, the bacteria were collected, and the genome was extracted. The specific steps are as follows:
[0039] The cells were resuspended with 500 ul of 50 mM Tris-HCl (pH 8.0) and 10 ul of 0.4 MEDTA, incubated at 37° C. for 20 minutes, and then 10 ul of 10 mg / ml lysozyme was added to continue the incubation for 20 minutes. Then add 3ul of 20mg / ml proteinase K, 15ul of 10% SDS, incubate at 50°C for 2 hours, then add 3ul of 10mg / ml RNase, and incubate at 65°C for 30 minutes. Add an equal volume of phenol for extraction, take the supernatant, and then extract twice with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), take the supernatant, and then extract with an equal volume of ether to remove residual phenol. Precipitate DNA with 2 times the volume of ethanol in the supernatant, roll out the DNA with glass wool and wash the DNA with 70% ethanol, and finally r...
Embodiment 2
[0040] Example 2: sequence deciphering
[0041] The genomes of Hafnia alvei G5902, G5903, G5904, and G5906 standard strains were extracted, and the whole genome sequence of each standard genome of Hafnia alvei was obtained by Solexapair-end sequencing technology to obtain the sequence of the O serotype. Blast and PSI-Blast for sequence comparison, TMHMMServer2.0program for transmembrane structure prediction, ClustalWprogram for sequence alignment and screening of conserved and specific gene fragments, and finally obtained the O antigen gene cluster sequence and decipherment of each serotype of Hafnia alvei result.
Embodiment 3
[0042] Example 3 : Primer design
[0043] According to the deciphering of the gene cluster and the results of database comparison, it was found that the wzy and wzx genes are specific genes for the O antigen of Hafnia alvei, so the specific segment of the gene was selected to design specific primers. Since wzy is more specific, the wzy gene is mainly used as the target gene.
[0044] Primer design is a core part of this invention. Primers were designed according to the specific genes described in the literature. The two genes wzx and wzy are relatively specific genes in the O antigen gene cluster of Hafnia alvei, which can be used as target genes for serotype identification. The above-mentioned genes were introduced into PrimerPremier5 for primer design. The length of the primers should preferably be between 18 and 24 bp, and the Tm value should be between 53 and 58°C. A pair of primers are designed for each gene, with a single target band.
[0045] After the primers a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com