Cytomembrane positioning signal peptide and coding sequence and application thereof
A technology for locating signals and cell membranes, applied in the field of genetic engineering, can solve problems such as difficult to treat diseases, difficult to meet the needs of breeding development, etc., and achieve the effect of high-efficiency drug delivery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Identification of Transit Signal Peptide
[0034] 1. Construction of recombinant plasmids
[0035] 1. DNA template preparation: Genomic DNA of Edwardsiella tarda LSE40 was extracted.
[0036]2.PCR amplification: with the DNA of step 1 as template, 108For(AT CATATG CCTTGGATGCCACGATTGC, the underline is the NdeI restriction site) and 108Rev( GAATTC TCGGCCTTGGCCTTTTGCTCGGCCT, the underline is the EcoRI restriction site) for PCR amplification to obtain a PCR product with a size of 324bp. The PCR amplification system (50μl) is: 10×buffer5μl, MgCl 2 (25mM) 5μl, dNTPmixture (10mMeach) 1μl, PfuTaqpolymerase (5U / μl) 0.5μl, 108For (10μM) 1μl, 108Rev (10μM) 1μl, DNA template 1μl, the rest add ddH 2 O. The PCR amplification conditions were: 94°C for 5 minutes; 35 cycles of 94°C for 30s, 55°C for 30s, and 72°C for 30s; 72°C for 10 minutes.
[0037] 3. Digestion: the PCR product in step 2 was double-digested with restriction endonucleases NdeI and EcoRI (37° C. for ...
Embodiment 2
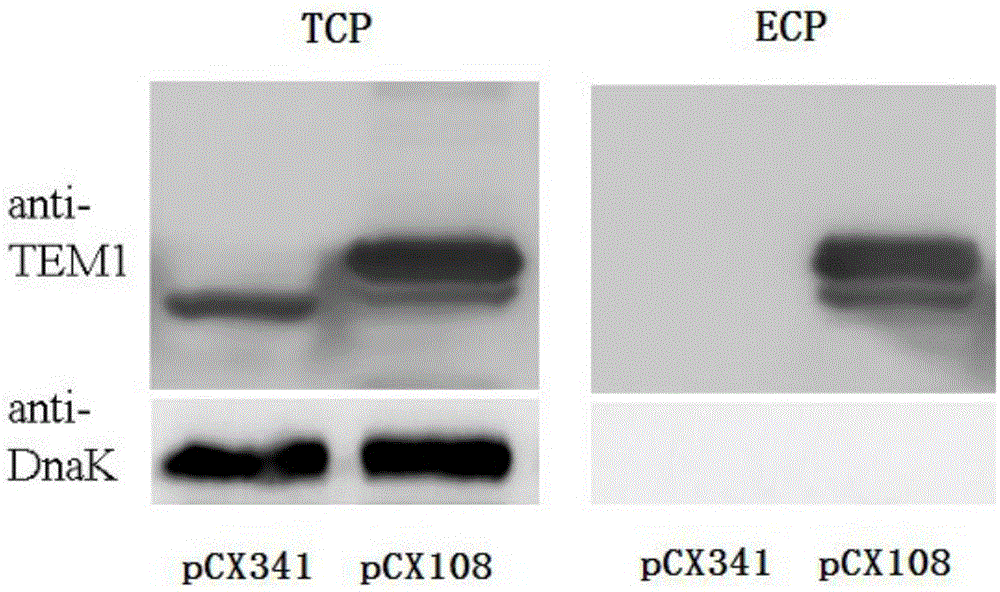
[0073] Implementation 2 Localization detection of fusion protein in flounder cells
[0074]1. Cultivation of recombinant bacteria: Inoculate LSE40pCX341 and LSE40pCX108 in TSB medium respectively, culture on a shaking table at 25°C for 18 hours, and transfer the bacterial solution to fresh TSB medium at a ratio of 1:20 (v:v) After culturing for 5 hours, take the bacterial solution and centrifuge at 5000g for 10 minutes at 4°C to collect the bacterial cells, wash three times with PBS and resuspend, adjust the bacterial concentration to 10 7 cells / ml.
[0075] 2. Infected flounder: buy flounder (Paralichthysolivaceus) from a farm and raise them temporarily in the aquarium for a week, then take 3 fishes, take internal organs for bacterial isolation, and use them for experiments after testing that there is no bacterial infection. Divide the flounder into two groups, 20 tails / group, respectively take the LSE40pCX341 and LSE40pCX108 bacterial solutions in step 1 and inject flounder...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com