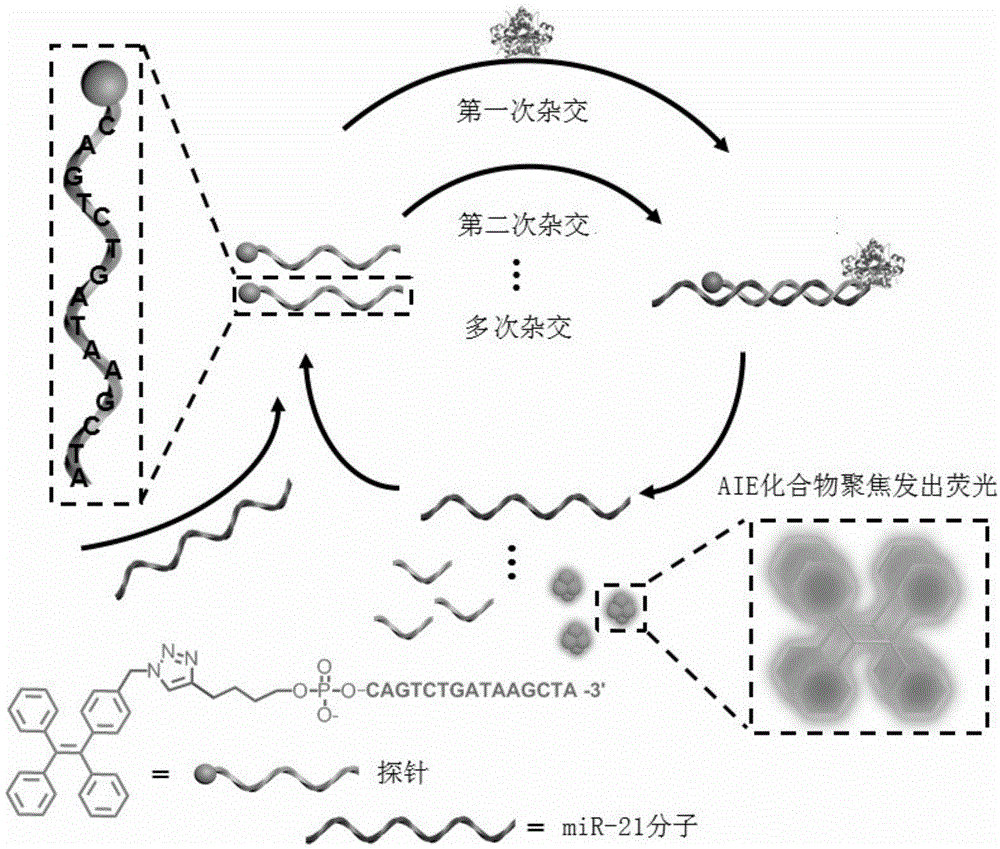
MicroRNA detection method
A detection method, a technology of RNA recombination, which can be applied to DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc. The effect of improving reaction sensitivity, reducing cost, and simple synthesis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] According to the sequence of miR-21 (SEQ NO:2), the probe molecule with DNA sequence such as SEQ NO:1 is designed, and the alkynyl group is modified at the 5' end, and 2-(4-(benzyl azide)phenyl- 1,1,2-Triphenylethylene reacts under the catalysis of cuprous bromide and sodium ascorbate, and the synthetic route is as follows:
[0036]
[0037] In the reaction, compound of formula II: compound of formula I: cuprous bromide: sodium ascorbate=1:10:11.5:23 (molar ratio); reaction condition: anaerobic environment, temperature is 25 ℃~30 ℃, stirs 24 hours or so. The reaction product was purified by reverse-phase high-performance liquid chromatography, and finally characterized by mass spectrometry to obtain the probe molecule with the molecular structure shown in formula III.
Embodiment 2
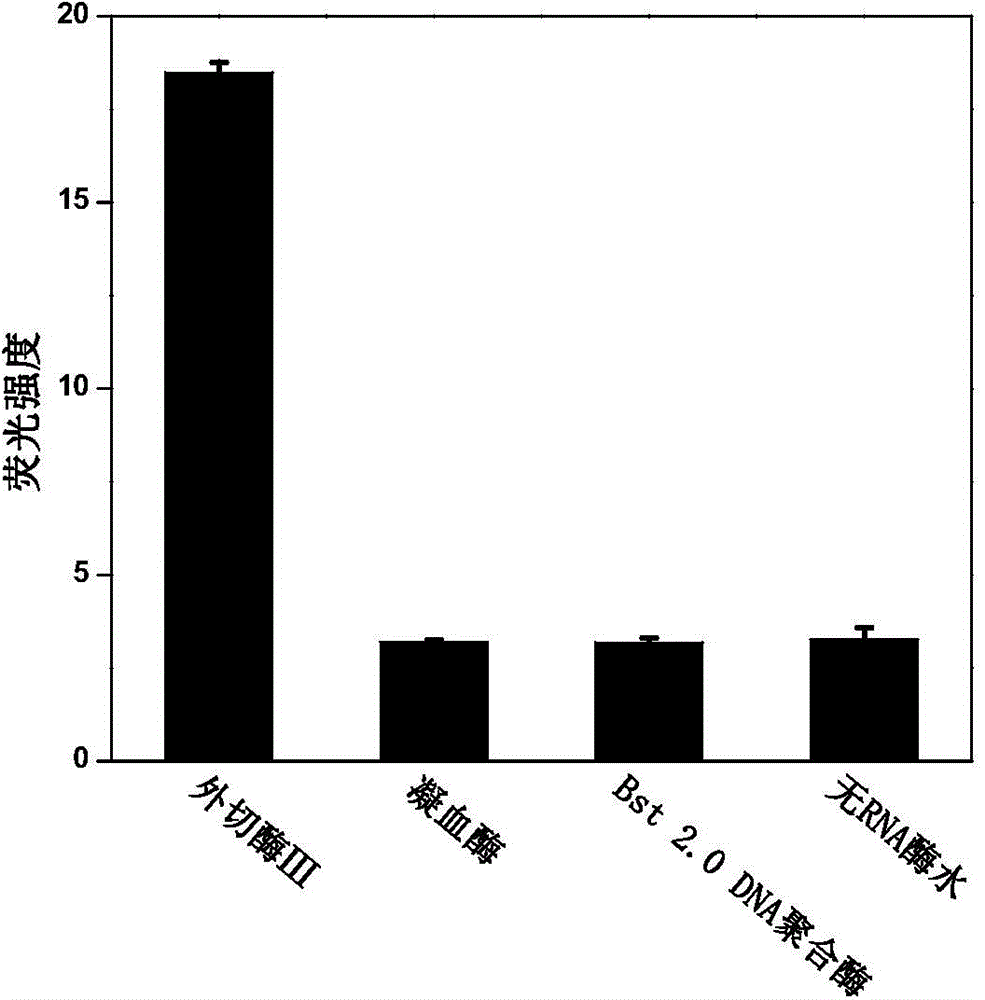
[0039] Step 1: Using the synthesized miR-21 as the test substance, prepare a 100nM solution. Add 23 μL RNA-free water, 5 μL NaCl solution (500 mM), 5 μL probe molecule (100 μM), 0.5 μL RNA recombinase inhibitor (40 U / μL), 5 μL miR-21 (100 nM), 5 μL into a 1.5 mL centrifuge tube, respectively. Graphene solution, 5 μL 10×DNA exonuclease III buffer, react in 37°C water bath for 30 minutes;
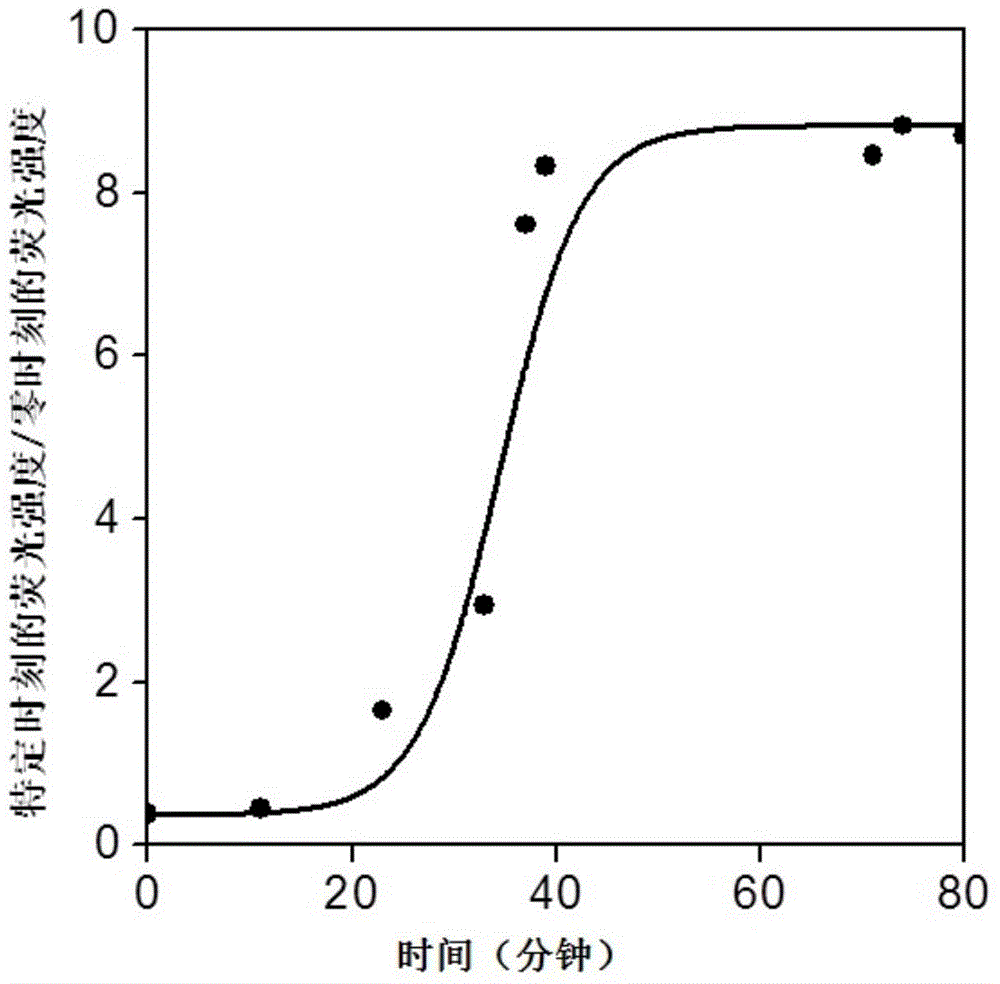
[0040] Step 2: Measure the fluorescence value immediately after adding 1.5 μL of DNA exonuclease III (200 U / μL); then measure the fluorescence value every 3 minutes at a temperature of 37°C, and obtain the curve of fluorescence intensity changing with time as shown in figure 2 , the measurement conditions of the fluorescence spectrum are: the excitation slit and the emission slit are both 5nm, the voltage is 600v, the excitation wavelength is 350nm, the emission scanning range is 400-600nm, and a 50μL quartz cuvette is used for measurement.
Embodiment 3
[0042] Repeat Example 1 with the same steps as described, the difference is that the concentration of miR-21 in Step 1 is 100aM (10 -16 mol / L); the reaction temperature in step 2 was plus 4°C, and the fluorescence intensity was measured after 3 days.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com