siRNA for inhibiting expression of FABP4 gene and application of sirna
A FABP4, gene expression technology, applied in the direction of DNA / RNA fragments, applications, gene therapy, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1 Synthesis of siRNA that inhibits FABP4 gene expression
[0040]Based on the known sequence of FABP4 in the GeneBank gene bank, the present invention designs three pairs of siRNA sequences according to the principle of siRNA design. Through cytological intervention, fluorescence localization and RT-PCR gene detection, it is verified that the following sequences can significantly inhibit FABP4 gene expression. The three pairs of siRNA sequences designed by the present invention are respectively (commissioned by genepharma biological company to synthesize):
[0041] siRNA-309:
[0042] Sense strand: 5'-GAGCAUCAUAACCCUAGAUtt-3'
[0043] Antisense strand: 5'-AUCUAGGGUUAUGAUGCUCtt-3'
[0044] siRNA-368:
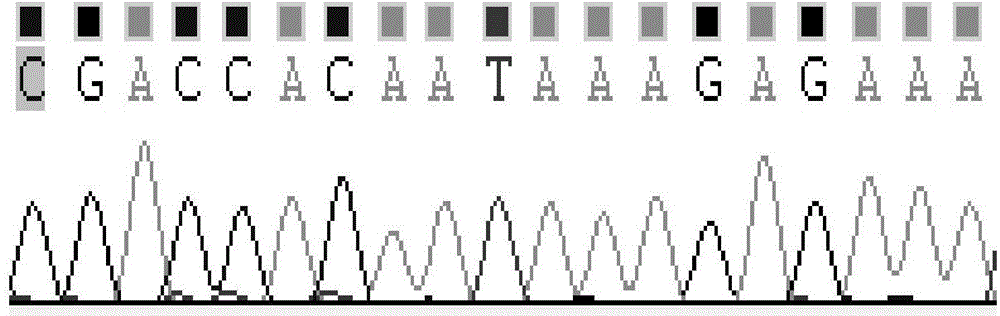
[0045] Sense strand: 5'-CGACCACAAUAAAGAGAAAtt-3'
[0046] Antisense strand: 5'-UUUCUCUUUAUUGUGGUCGtt-3'
[0047] siRNA-432:
[0048] Sense strand: 5'-CGACCACAAUAAAGAGAAAtt-3'
[0049] Antisense strand: 5'-UUUCUCUUUAUUGUGGUCGtt-3'
[0050] The negative cont...
Embodiment 2
[0055] Example 2 Cell culture and transfection inhibit the expression test of FABP4 gene
[0056] 1. Cell culture
[0057] (1) Recovery of 3T3-L1 preadipocyte line
[0058] Take out the cell line from liquid nitrogen, put it in a 37°C water bath immediately, and revive it by rapid shaking for about 1.5-2min. Add the recovered cells into a 50ml centrifuge tube containing 5ml of culture medium, and centrifuge at room temperature for 4min at 900rpm. Pour off the supernatant, add 5ml of culture medium, and observe the cell morphology and number under a microscope. placed in 5% CO 2 Cultured in a constant temperature incubator at 37°C, and the medium was changed after 24 hours. The 3T3-L1 preadipocyte cell line used in the present invention was kindly donated by the Laboratory of Endocrinology Department of Shanghai Ruijin Hospital.
[0059] (2) Culture and passage of 3T3-L1 preadipocyte line
[0060] Observe under a microscope that the cells have adhered to the wall, appear ...
Embodiment 3
[0067] Embodiment 3 RT-PCR experiment detects FABP4 gene expression
[0068] After siRNA transfection into cells, in 5% CO 2 Incubate for 48 hours at 37°C in an incubator. Total cellular RNA was extracted by Trizol method and reverse transcribed into cDNA.
[0069] Entrust Shanghai Bioengineering Company to synthesize FABP4 primers, primer sequence:
[0070] FABP4-F: CATGGCCAAGCCCAACAT
[0071] FABP4-R: CGCCCAGTTTGAAGGAAATC
[0072] The RT-PCR system is as follows:
[0073] Element
volume
2×SYBR Green Mix
10ul
FABP4 Primer Mix
1ul
cDNA
2ul
wxya 2 o
7ul
total capacity
20ul
[0074] Aliquot to AXYGEN PCR8-connected tubes, and centrifuge briefly in a microcentrifuge to mix. The above samples were put into the Eppendorf Realplex fluorescent quantitative PCR instrument, and the SYBR Green method fluorescent quantitative PCR was used to analyze the expression of each gene. The PCR program was set as...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com