Molecular detection kit for determining resistance level of Escherichia coli to quinolone drugs based on high resolution melting curve
A quinolone, Escherichia coli technology, applied in the field of molecular detection kits, can solve the problems of reducing bacterial sensitivity, time-consuming and the like, and achieve the effect of accurate judgment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Gyrase A The nucleotide sequences of the primers and probes used in the FRET-qPCR detection method are as follows:
[0039] Upstream primer: 5’- ctttacgccatgaacgtactaggc -3’
[0040] Downstream primer: 5’- tttccgtaccgtcatagttatcaac -3’
[0041] Probe-1: 5’-ggggatggtatttaccgattacgtcaccaacgaca-6-FAM-3’
[0042] Probe-2: 5'-LCRED640-acgatcgtgtgtcataaaccgccgagtcacca-phospho-3'.
Embodiment 2
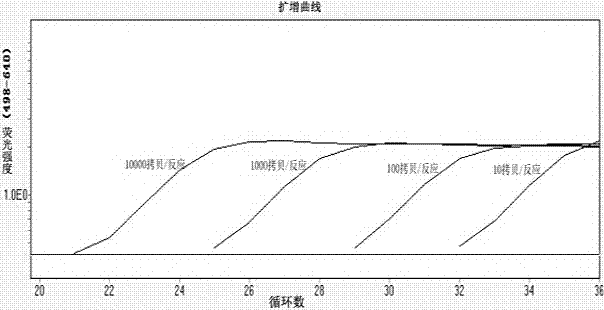
[0044] Preparation of standard quantitative reagents for PCR: synthesized by IDT Gyrase A The nucleic acid sequence of a gene covering the region amplified by PCR. Based on the molecular weight and absolute weight of the compound and PicoGreen's DNA quantification technology, the amount of DNA contained in the compound is calculated. Gyrase A Gene copy number. Subsequently, the compound was diluted to prepare dilution reagents containing 10000 copies, 1000 copies, 100 copies and 10 copies of the target gene per 10 μl of the compound, which were used as standard quantitative reagents for PCR.
[0045] Prepare the DNA template of the sample to be tested: nucleic acid extraction of Escherichia coli: tissues (semen, milk) known to contain E. 10 E. 0.1 (containing 10 mM Tris-HCl, 0.1 mM EDTA, pH 8.5) as the amplification template for PCR. Nucleic acid extraction of Escherichia coli: For organs known to contain Escherichia coli (liver, lung, intestine), weigh about 20 mg of the...
Embodiment 3
[0047] PCR amplification system: 20μl amplification system, including 10μl sample DNA template or quantitative standard reagent, 1xPCR buffer, 1μM upstream primer-1, 1μM downstream primer, 0.2μM 6-FAM probe, 0.2μM LCRed640 probe Needle, 2 units of commercial Taq enzyme, 200 μM dNTPs.
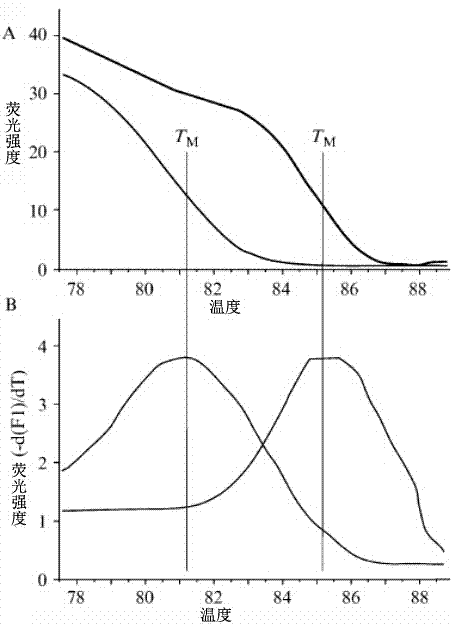
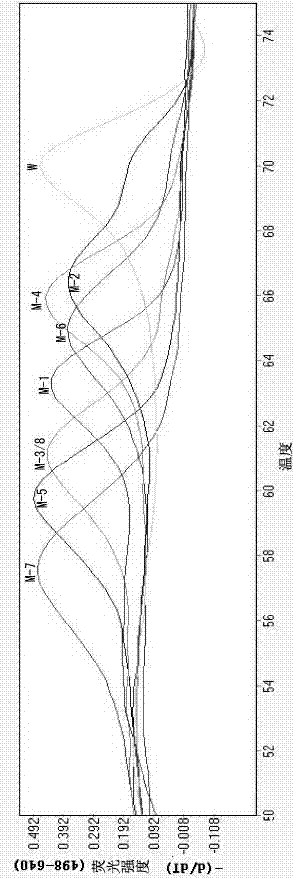
[0048] PCR amplification cycle parameters: PCR amplification includes 40 fluorescence acquisition cycles: 40 x 10 sec 95°C, 15 sec 56°C, 15 sec 72°C. After the PCR amplification, a high-resolution melting curve analysis is set up: the temperature is gradually increased from 45 °C to 85 °C, with an increment of 0.11 °C per second, and the change of fluorescence intensity is continuously monitored. Data analysis is to analyze the ratio of fluorescence intensity at 640 nm:530 nm (F4 / F1). The first derived value of F4 / F1 (-d(F4 / F1) / dt) is read as the melting temperature of the DNA.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com