Nucleotide sequence and detection method for detecting purity of Jinyou #401 cucumber hybrid seed
A technology of hybrid seed purity and nucleotide sequence, applied in the field of DNA sequence, can solve problems such as long identification period, fuzzy identification standard, and certain degree of deviation in phenotypic characteristics, and achieve fast and accurate identification results, great application value, and saving human effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] The method for detecting the purity of Jinyou No. 401 cucumber hybrid seeds comprises the steps:
[0023] Genomic DNA was extracted from the root tip of Jinyou 401 cucumber hybrid seeds 36 hours after germination; detailed steps for DNA extraction by CTAB method:
[0024] Add a 1-2cm long root tip to a 1.5mL centrifuge tube; use tweezers to place the root tip at the bottom of the centrifuge tube, add 2 steel balls to each centrifuge tube; add 400 μL CTAB buffer, cover the centrifuge tube cap, and place Place the centrifuge tube on a special 45-hole plate for the grinding machine; clamp the 45-hole plate on the grinder, and grind for 30 seconds; take the centrifuge tube out of the 45-hole plate, place it on a 24-hole rack, and heat it in a water bath at 65°C for 1 hour. Invert the centrifuge tube up and down for 3-5 times within 10 minutes to ensure that the CTAB buffer can fully extract the DNA in the tissue; add 400 μL 24:1 chloroform / isoamyl alcohol, and mix up an...
Embodiment 2
[0030] The method for detecting the purity of Jinyou No. 401 cucumber hybrid seeds is characterized in that it is carried out according to the following steps:
[0031] (1) Genomic DNA was extracted from cucumber hybrid seeds of Jinyou 401;
[0032] (2) PCR amplification: add Jinyou 401 Cucumber hybrid seed genomic DNA 20ng into a thin-walled tube dedicated to PCR amplification, then add 25ng of upstream primers, 25ng of downstream primers, 1 times Mg-containing 2+ PCR buffer, dNTP 2mmol, Taq DNA polymerase 1 unit, add sterile double distilled water to 10μl; put the special thin-walled tube for PCR amplification into the PCR instrument for amplification, the amplification conditions are: 94 ℃ pre-denaturation for 180 seconds ; Denaturation at 94°C for 30 seconds, annealing at 50-55°C for 30 seconds, extension at 72°C for 60 seconds, 30 cycles; extension at 72°C for 7 minutes, the amplification is complete;
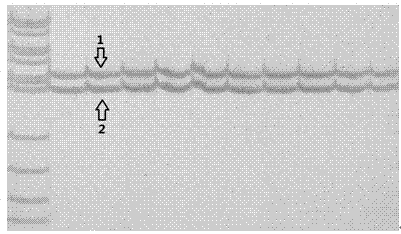
[0033](3) Gel electrophoresis analysis of PCR amplification products...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com