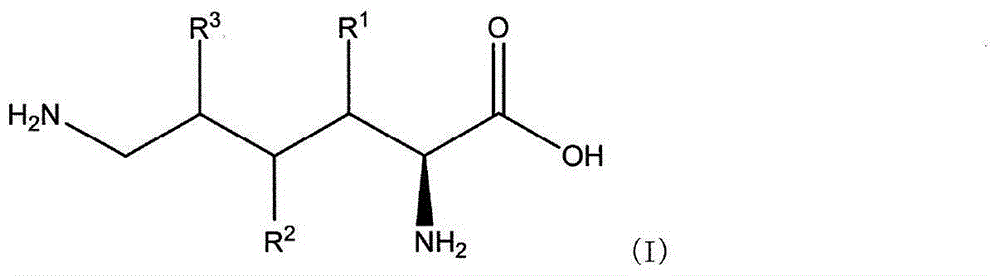
Production method for l-lysine hydroxylase and hydroxy-l-lysine using same, and production method for hydroxy-l-pipecolic acid
一种赖氨酸羟化酶、赖氨酸羟化酶活性的技术,应用在生物化学设备和方法、氧化还原酶、酶等方向,能够解决尚未报道L-赖氨酸等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0184] Cloning of 2-oxoglutarate-dependent L-lysine hydroxylase gene
[0185] Based on the gene sequence (hyl-1, sequence No. 1), designed and synthesized the full-length primers hyl1_F (SEQ ID NO: 13) and hyl1_R (SEQ ID NO: 14) for amplifying the hyl-1 gene. Using the chromosomal DNA of Flavobacterium johnii as a template, PCR reaction was carried out according to conventional methods to obtain a DNA fragment of about 1.0 kbp.
[0186] Similarly, for Kineococcus radiotolerans NBRC101839 strain, Chitinophaga pinensis NBRC15968 strain, Chryseobacterium gleum NBRC15054 strain, and Niastella koreansis NBRC106392 strain The source of VioC homologues are Hyl-2 (GenBank Accession No. ABS05421, Sequence No. 4), Hyl-3 (GenBank Accession No. ACU60313, Sequence No. 6), Hyl-4 (GenBank Accession No. EFK34737, Sequence No. No. 8), Hyl-5 (GenBank Accession No. AEV99100, Sequence No. 10). Based on the gene sequences encoding each enzyme (hyl-2 (SEQ ID NO: 3), hyl-3 (SEQ ID NO: 5), h...
Embodiment 2
[0191] Activity Confirmation of 2-oxoglutarate-dependent L-lysine Hydroxylase Based on Resting Cell Response
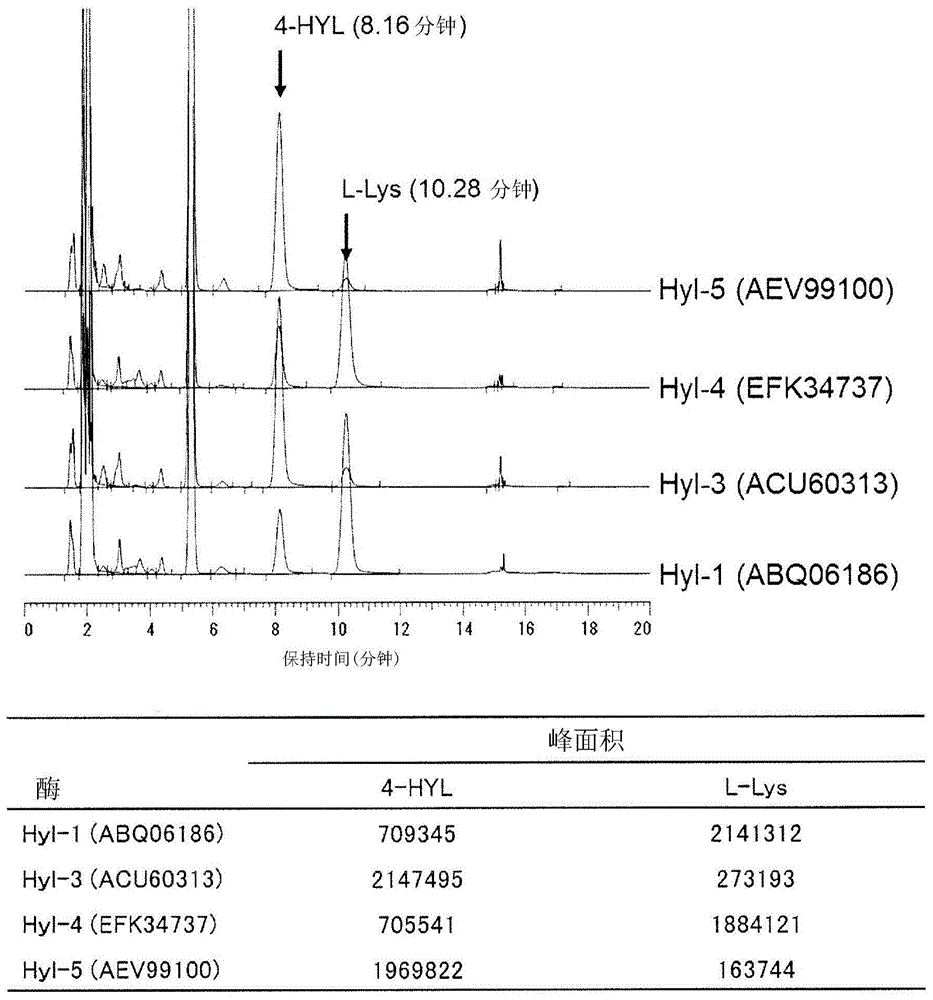
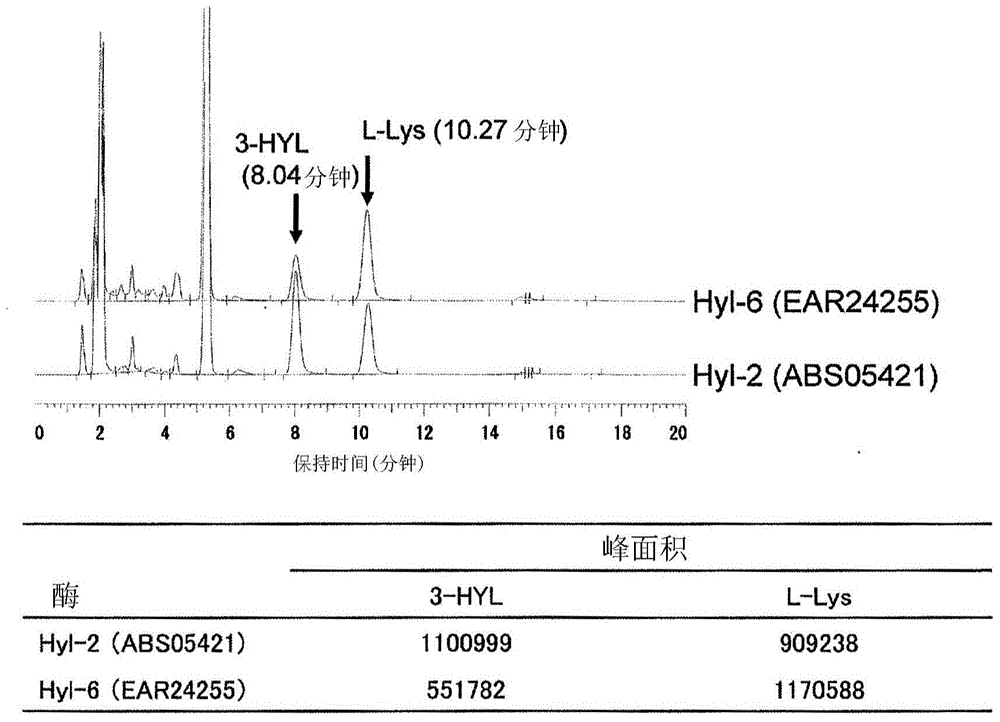
[0192] Mix 5mM L-lysine, 10mM 2-oxoglutaric acid, 1mM L-ascorbic acid, 0.1mM ferrous sulfate, and the recombinant Escherichia coli obtained by the method according to Example 1 in a plastic tube, according to the turbidity OD 600 =10 way to mix the reaction solution. 0.5 ml of the prepared mixed solution was reacted at 30° C. and pH 7.0 for 3 hours. The reaction products were derivatized with 1-fluoro-2,4-dinitrophenyl-5-L-alaninamide (FDAA) and analyzed by HPLC. As a result, such as figure 1 , figure 2 As shown, it was confirmed that BL21(DE3) / pEHYL2 and BL21(DE3) / pJHYL6 produced consistent compounds when the retention time of the 3-hydroxylysine standard substance was 8.04 minutes. In addition, it was confirmed that BL21(DE3) / pEHYL1, BL21(DE3) / pEHYL3, BL21(DE3) / pEHYL4, and BL21(DE3) / pEHYL5 produced consistent 4-hydroxylysine standard substances at a rete...
Embodiment 3
[0196] Synthesis of (2S,3S)-3-hydroxylysine
[0197] Mix 35mL of 1M potassium phosphate buffer (pH7.0), 304mL of desalted water, 1.28g of L-lysine hydrochloride, 2.05g of 2-ketoglutarate, 0.14g of sodium L-ascorbate, and sulfuric acid in a 1L fermenter. 0.02g of ferrous iron, 0.35g of Adekanol LG109, and 8g of wet bacteria of recombinant Escherichia coli BL21(DE3) / pEHYL2 obtained according to the method of Example 1, at 30°C, pH7.0, agitation number 500rpm, air ventilation Under the condition of volume 2.0vvm, react for 17 hours. The end of the reaction was judged by confirming that the peak of L-lysine disappeared by HPLC analysis. By centrifugation and microfiltration, bacteria and bacteria residues were removed from the liquid after the reaction to obtain 390 g of filtrate.
[0198] In addition, the analysis conditions of L-lysine by HPLC are as follows.
[0199] Column: SUMICHIRAL OA-6100 (4.6mm×250mm) manufactured by Sumika Analytical Center, mobile phase: 1mM ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com