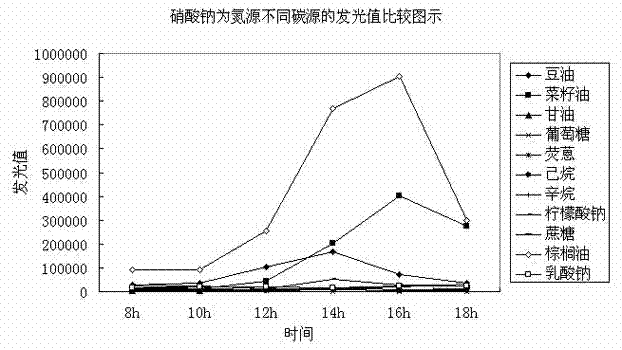
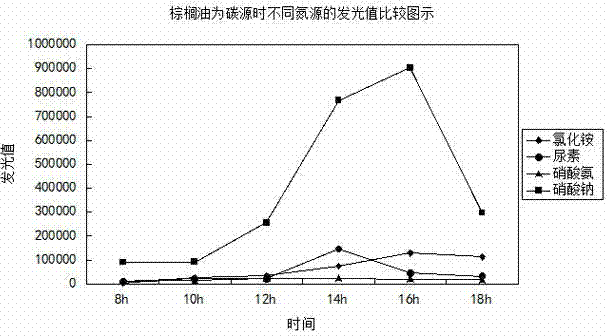
Method for rapidly and efficiently screening rhamnolipid producing bacteria nutrition system
A technology for producing rhamnolipids and bacteria, which is applied in the direction of microorganism-based methods, biochemical equipment and methods, and microbial measurement/inspection, and can solve problems such as low yield, high production cost, and inability to achieve a single product
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] A method for rapidly and efficiently screening the nutritional system of rhamnolipid-producing bacteria, comprising the following steps:
[0038] 1) Synthesize the rhamnolipid production gene in a related gene synthesis company wxya Primers for gene amplification by PCR, synthesized wxya Gene fragments, using genetic engineering cutting and splicing technology, will wxya The gene fragment was ligated to the pMS402 plasmid lux Before the luminescent gene, construct a recombinant plasmid, then transfer the above recombinant plasmid into Pseudomonas aeruginosa DN1, spread the recombinant strain on an LB solid plate containing 300 mg / LTmp, screen out positive clones, and name the strain as rhlAB-lux For bacterial strains, positive monoclonal colonies were selected and enriched on solid LB medium, and stored at 4°C for later use;
[0039] 2) Take refrigerated rhlAB-lux The strains were activated on LB solid medium containing 300mg / LTmp (trimethoprim) for 24h, single c...
Embodiment 2
[0047] A method for rapidly and efficiently screening the nutritional system of rhamnolipid-producing bacteria, comprising the following steps:
[0048] 1) Synthesize the rhamnolipid production gene in a related gene synthesis company wxya Primers for gene amplification by PCR, synthesized wxya Gene fragments, using genetic engineering cutting and splicing technology, will wxya The gene fragment was ligated to the pMS402 plasmid lux Before the luminescent gene, construct a recombinant plasmid, then introduce the above recombinant plasmid into Pseudomonas aeruginosa PAO1, spread the recombinant strain on an LB solid plate containing 200 mg / LTmp, screen out positive clones, and name the strain as PAO1-lux For bacterial strains, positive monoclonal colonies were selected and enriched on solid LB medium, and stored at 4°C for later use;
[0049] 2) Take refrigerated PAO1-lux Bacterial strains were activated on LB solid medium containing 200mg / LTmp (trimethoprim) for 24h, si...
PUM
| Property | Measurement | Unit |
|---|---|---|
| surface tension | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com