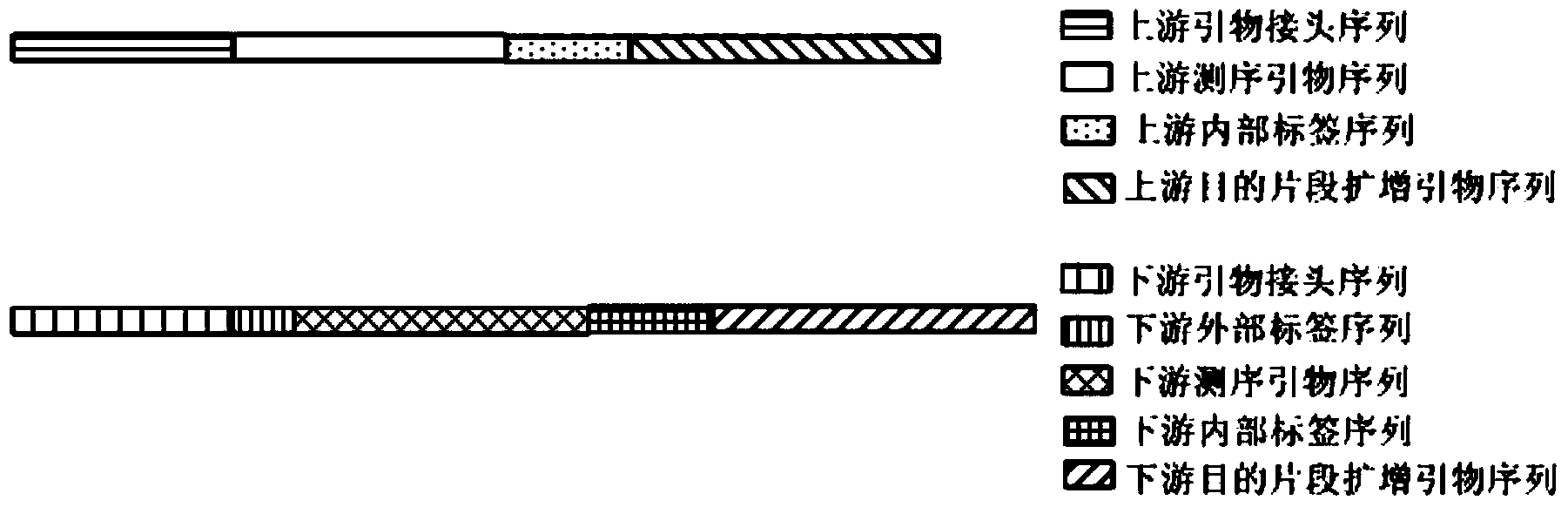
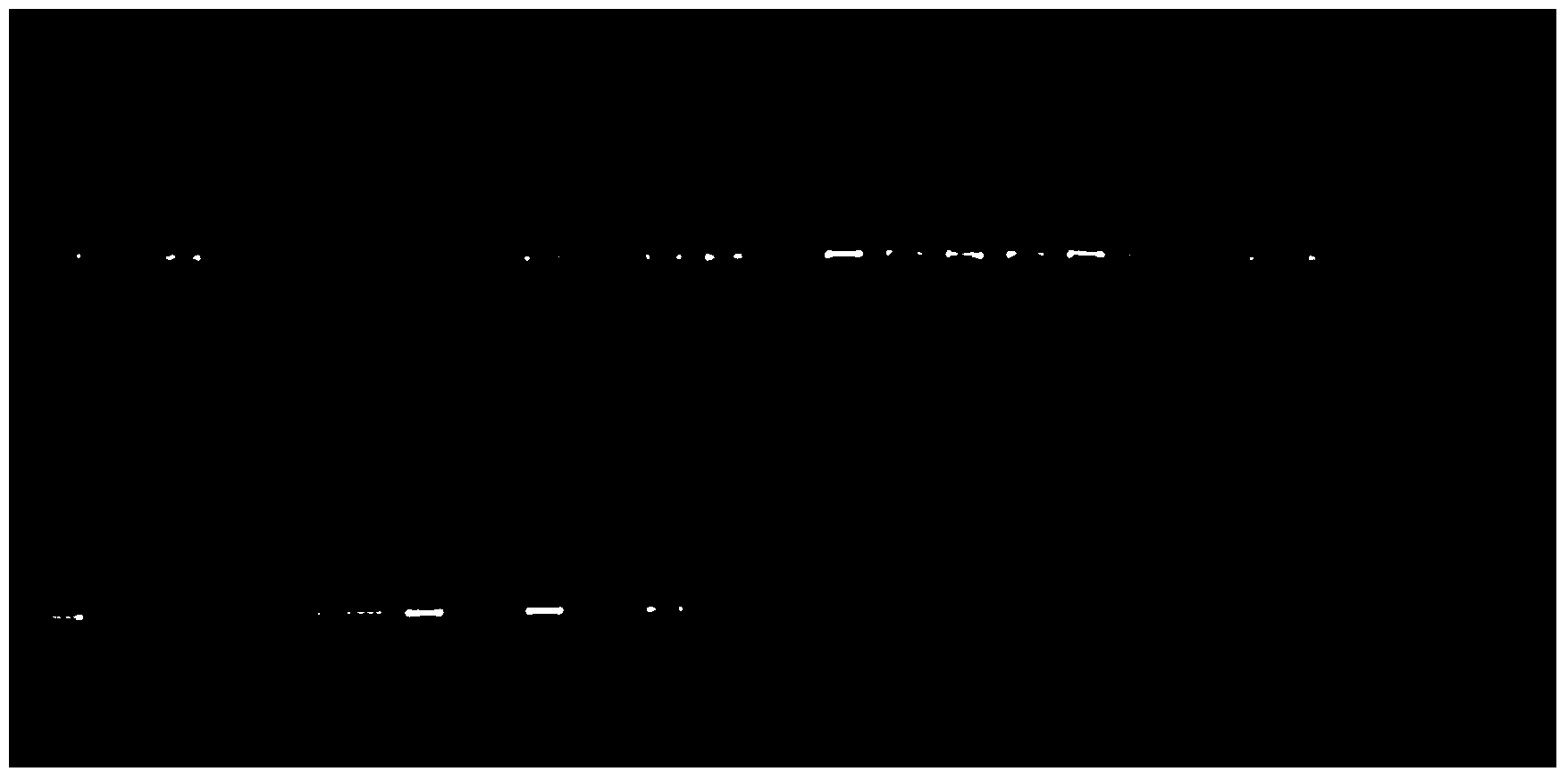Primer applicable to amplicon sequencing library construction, construction method, amplicon library and kit comprising amplicon library
A technology of amplicon library and amplification primer, which is applied in libraries, chemical libraries, nucleotide libraries, etc., can solve the problems of overestimating microbial diversity and low accuracy of sequencing data, and increase the number of mixed samples , Improve throughput and improve accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] A method for constructing a 16S rDNA v4 region amplicon sequencing library applied to Illumina MiSeq sequencing, comprising the following experimental steps:
[0048] (1) According to the sample 16S rDNA DNA concentration, use sterile water to dilute the sample to 1ng / μl. If there is a large amount of RNA contamination in the sample, it needs to be digested with RNase before dilution.
[0049] (2) PCR amplification. Use the synthesized primers shown in Table 1 to amplify according to the combinations shown in Table 2.
[0050] Table 1:
[0051] Numbering
Primer sequence
SEQ ID NO.1
AATGATACGGCGACCACCGAGATTCACTCTTTCCCTACACGACGCTCTTCCGATCTATCACGGTGCCAGCMGCCGCGGTAA
SEQ ID NO.2
AATGATACGGCGACCACCGAGATTCACTCTTTCCCTACACGACGCTCTTCCGATCTCGATGTGTGCCAGCMGCCGCGGTAA
SEQ ID NO.3
AATGATACGGCGACCACCGAGATTCACTACTCTTTCCCTACACGACGCTCTTCCGATCTTTAGGCGTGCCAGCMGCCGCGGTAA
SEQ ID NO.4
AATGATACGGCGACCACCGAGATTCACTCTTTCCCTACACGACGCTCTTCC...
Embodiment 2 and
Embodiment 2
[0069] Example 2 uses the same steps as Example 1, only the primers are different. The specific primer sequences are shown in Table 3, and the amplification results are shown in image 3 . image 3 The order of the samples from left to right is the order listed in Table 4.
[0070] table 3
[0071] Numbering
[0072] Table 4
[0073] Sample serial number
[0074] image 3 Among them, the size of the target fragment of the amplified 16S rDNA is about 300bp, and the total length of the upstream primer and downstream primer is about 380bp, excluding 19bp of the upstream primer and 20bp of the downstream primer of the target fragment. The icon size is also consistent with the expected size. From image 3 It can be seen from the figure that in the two-step amplification library construction method, the target fragment can be amplified well by having internal tag sequences at both ends of the target fragment amplification primer sequence. The linker sequ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com