Establishing method of cymbidium microsatellite labels, core fingerprint label database and kit
A technology of microsatellite marker and establishment method, which is applied in the field of fingerprint marker library of main species of Orchid, can solve the problems of large overall information of the genome, unknown, large manpower and material resources, etc., and achieves good versatility, good versatility and conservation. high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1 The preparation and screening method of SSR markers in the genus Orchid
[0033] The present embodiment is the preparation and screening method of SSR marker of Orchid, which mainly includes the following steps:
[0034] 1. Acquisition of Jianlan gene expression sequence:
[0035] Select the buds of Jianlan to extract total RNA, use the standard Solexa sequencing method to obtain transcriptome Reads, remove low-quality, carrier and non-coding sequences (such as rRNA, tRNA and miRNA), use Trinity splicing software to complete the sequence assembly, and finally obtain 101423 Jianlan The non-redundant Unigene sequence of .
[0036] 2. Retrieval of SSR sites in Jianlan transcriptome sequence:
[0037] Using MSATCOMMANDER to analyze the occurrence frequency of SSR. The minimum number of repeating motifs was set to 10 times for single nucleotides, 6 times for dinucleotides, 4 times for trinucleotides, tetranucleotides, pentanucleotides, and hexanucleotides, and a...
Embodiment 2
[0048] Embodiment 2: the universality test of blue genus mark
[0049] 1. Experimental materials:
[0050] A total of 48 materials from five species of Orchid (Chunlan, Cymbidium, Jianlan, Molan and Hanlan) were selected to test the generality of Jianlan SSR markers in Orchid plants. For specific material information, see Table 1.
[0051] 48 pieces of test material information for carrying out primer screening and genetic analysis in the present invention in table 1
[0052]
[0053] 2. Genome extraction and SSR primer amplification and screening
[0054] Select about 200mg of young leaves for each material, put them into a 2.0ml centrifuge tube, and put steel balls with a diameter of 0.5cm into them. After being quickly frozen in liquid nitrogen, shake them on a homogenizer for 35s to grind the tissues into powder. CTAB method for extraction. Prepare 8% polyacrylamide gel in the DYCZ-30 type electrophoresis tank (Beijing Liuyi Instrument Factory) for electrophoresis se...
Embodiment 3
[0061] Embodiment 3: the establishment of Orchid genus core marker library
[0062] 1. SSR primer amplification results statistics and polymorphism analysis
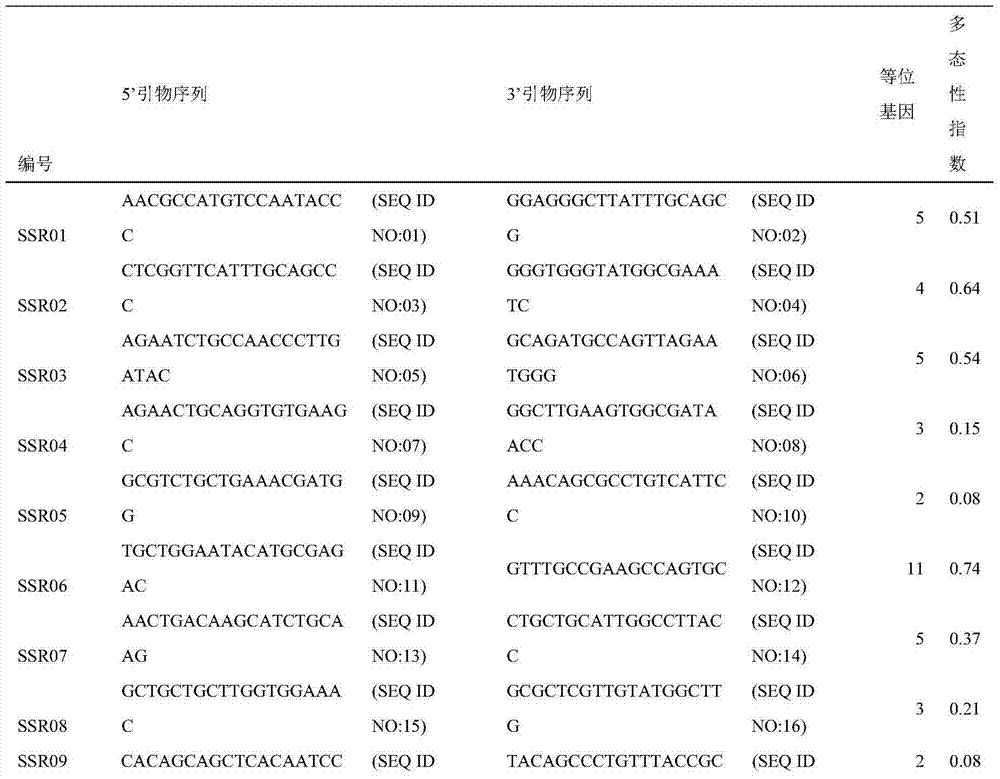
[0063] According to the electrophoresis results of step (2) in Example 2, the different genotypes are marked as 1, 2, 3, etc., and then input into Powermarker to count the polymorphisms of these markers. Among the 61 pairs of primers, 49 pairs had polymorphisms (see Table 2 for 49 markers), and a total of 216 genotypes were detected in 48 test materials, with an average of 3.7 genotypes per pair of primers. It shows that Jianlan SSR has high applicability in Orchid, and can be used to analyze the genetic diversity, phylogenetic relationship and genetic mapping of Orchid plants.
[0064] Table 2 49 orchid polymorphic marker sequences and related information in the present invention
[0065]
[0066]
[0067]
[0068] 2. Analysis of SSR primers' interspecies discrimination in Orchid:
[0069] For the above 49 p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com