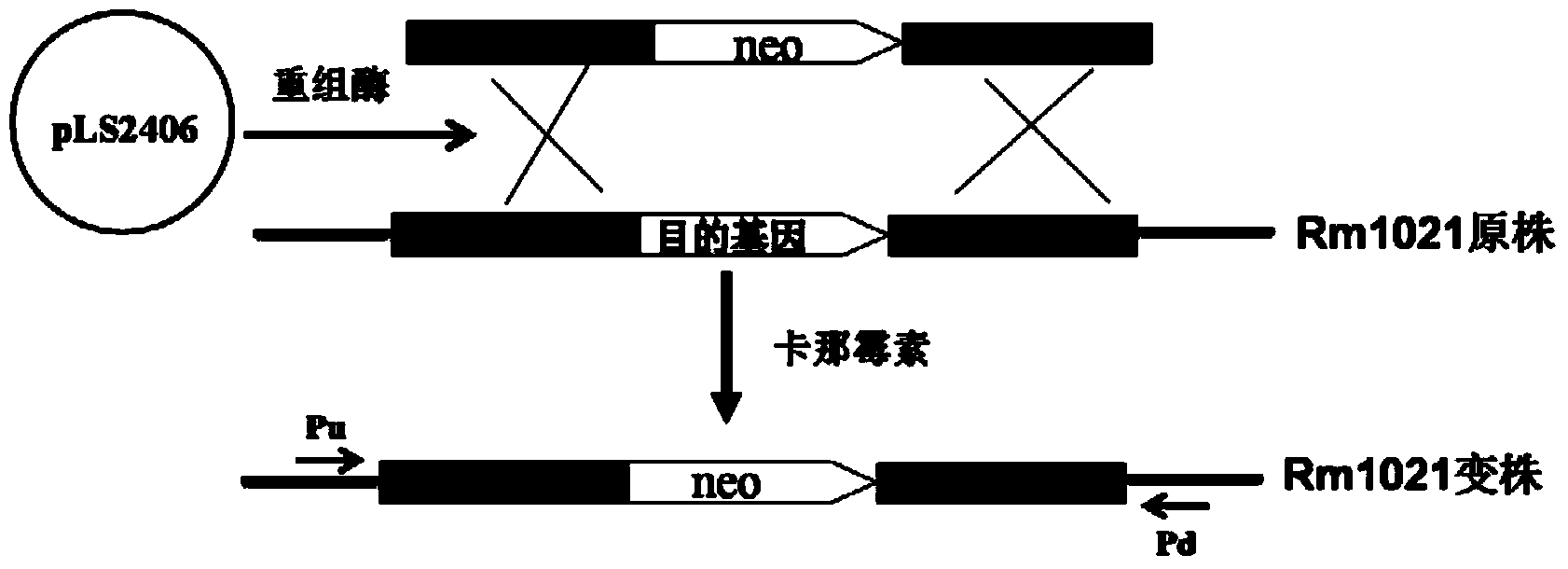
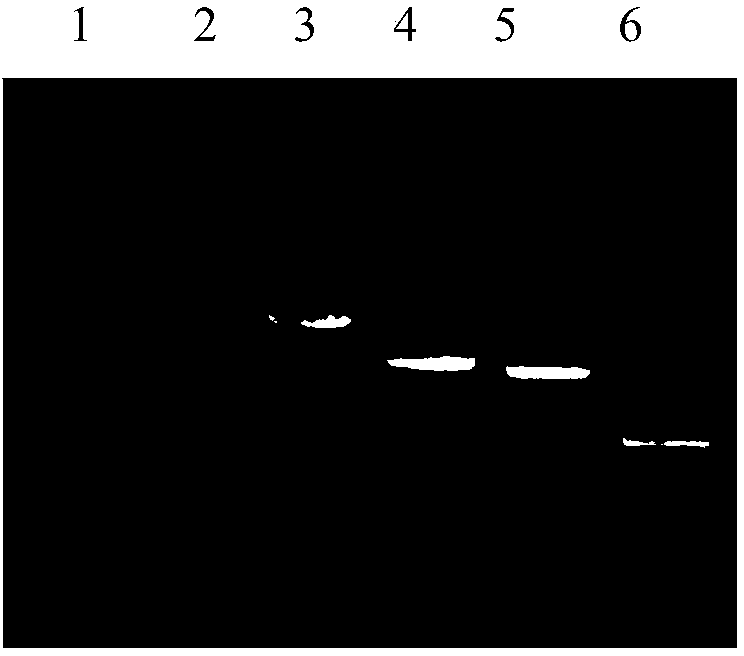
Re-engineering mediated sinorhizobium meliloti Rm1021 gene knockout method
A technology of Sinorhizobium, rm1021, applied in the field of genetic engineering, can solve the problems of cumbersome efficiency, time-consuming, highly inactive N-N health and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1. Construction of Recombinase Expression Plasmid
[0042] Design primer RN1:5'-GAA GAGCTCCATATG GATATTAATACTGAAACTG-3', (SEQ ID NO.1), the designed SacI and NdeI sites are underlined; RN2: 5'-GAA TCTAGA GTCATCGCCATTGCTCCCCAAATAC-3', (SEQ ID NO. 2), the designed XbaI site is underlined. Using lambda phage DNA (purchased from Shanghai Sangon Biological Co., Ltd.) as a template, RN1 and RN2 were PCR amplified to obtain a 1.9kb DNA fragment, digested with SacI and XbaI, and cloned into pBluescript II KS(-) digested with the same enzyme, After sequencing verification, the recombinant clone pNdRed was obtained. pNdRed was digested with NdeI and XbaI, and a 1.9kb DNA fragment was isolated, connected to the pSRKGm digested with the same enzyme, and 25 μg / ml gentamycin, IPTG and X-gal (5-bromo-4 chloro-3 -indole-β-D-galactoside) for screening. The white clone was picked and identified by plasmid digestion, and the recombinant clone pLS2405 was obtained. pKsacB wa...
Embodiment 2
[0043] Example 2. Knockout of the bacA gene on the S. meliloti Rm1021 genome
[0044] 1. Preparation of Rm1021 electroporation competent cells and transformation of pLS2406
[0045] Streak Rm1021 into LBMC solid medium from the cryopreservation tube at -80°C, and culture at 30°C for about 60 hours. Transfer a single colony to 3ml LBMC, shake culture at 30°C, 220rpm for about 36h, then transfer 2ml to 100ml LBMC, adjust the OD600 to about ~0.1, continue to culture until the OD600 is about 0.6, centrifuge, and discard the supernatant. The cell pellet was washed three times with ice-cold 10% glycerol, finally suspended in 200 μl of ice-cold 10% glycerol, and distributed in 50 μl. Add 100ng of pLS2406 to the competent cells, flick and mix well, transfer to a 1mm electric cup cooled on ice, and transform with 2.1kV electric shock, the electric transformation instrument is the Gene Pulser of Bio-Rad, USA II . After electrotransformation, add 1ml of SOC to suspend, and incubate at...
Embodiment 3
[0059] Example 3. Knockout of the exoR gene on the S. meliloti Rm1021 genome
[0060] The strategy of exoR gene knockout is the same as that of bacA. Design three pairs of primers R1341: 5'-TATCCTGACCAGCGGCAGTTC-3', (SEQ ID NO.13), R1342: 5'-GTGCGCGGAACCCCTATTTGTTCATAACAATCAGTTTCTTTCGTTC-3', (SEQ ID NO.14); R1343: 5'-GAACGAAAGAAACTGATTGTTATGAACAAATAGGGGTTCCGCGCAC-3' (SEQ ID NO. 15), R1344: 5'-GTCAGGCCGCACGGGACCTCATCCTTAGTTCCTATTCCGAAGTTC-3', (SEQ ID NO. 16); R134: 5'-GAACTTCGGAATAGGAACTAAGGATGAGGTCCCGTGCGGCCTGAC-3', (SEQ ID NO. 17), R1346: 5'-GGAAAAGACCCCCTA -3', (SEQ ID NO. 18). Wherein R1342 and R1343 and R1344 and R1345 are reverse complementary sequences, which are the overlapping sequence parts between the templates of the second step OE-PCR.
[0061] Using Rm1021 genomic DNA as a template, R1341 and R1342 PCR, R1325 and R1326 were used to amplify the exoR gene upstream and downstream respectively 0.5 kb; using pLS1918 as a template, R1343 and R1344 PCR amplified a 1.0 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com