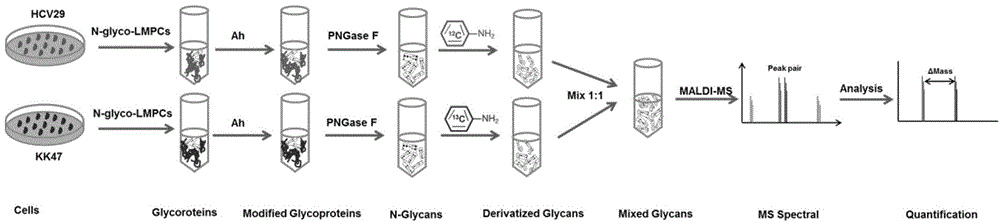
Method for relatively quantitatively analyzing carbohydrate chain in two-end labeling manner
A quantitative analysis and sugar chain technology, applied in the analysis of materials, material analysis by electromagnetic means, measurement devices, etc., can solve the problems of loss of sialic acid information, masking of sugar chain structure information, etc., to improve the comprehensiveness and quantitative accuracy. sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The preparation of embodiment 1 sample protein
[0029] Preparation of cell protein: Extract cell protein with a cell protein reagent extraction kit (such as T-PER, RIPA), use as little lysate as possible to ensure a protein concentration of 2-10 mg / mL, and centrifuge at 14,000g for 5 minutes to remove cell residues. The protein concentrations of the two cell extracts were determined with a BCA kit (guaranteed at least three repetitions).
[0030]Serum protein preparation: Serum samples were centrifuged at 12,000g for 15 minutes, and the middle liquid part was taken. Add to a 10KD ultrafiltration tube, place the ultrafiltration tube in a matching centrifuge tube, centrifuge at 12,000g for 15min, add 400μl of 40mM NH 4 HCO 3 , centrifuge again, and repeat. Invert the filter membrane into a new centrifuge tube, centrifuge at 9000g for 3min, collect the isolated protein (about 50μl), quantify it with the Bradford method, and finally adjust the volume to 2mg / ml.
Embodiment 2
[0031] Example 2 Labeling and analysis of sugar chain ends
[0032] (1) Acethydrazide modification and release of sugar chain sialic acid
[0033] Take 2mg of the protein extracted in Example 1 and add it to a 10KD ultrafiltration membrane, centrifuge at 14000g to concentrate to about 50μL, add 300μL of 8M urea, mix well, and centrifuge at 14000g for 15min. Add 200 μL of 8M urea solution to the ultrafiltration tube, and centrifuge at 14000 g for 15 min. Discard the effluent in the collection tube, add 150 μL of 10 mM DTT (dithiothreitol) and mix well, incubate at 56 ° C for 45 min, centrifuge at 14000 g for 10 min, add 150 μL of 20 mM IAM (iodoacetamide) and mix well, dark Incubate at 20min, centrifuge at 14000g for 10min. Add 150 μL of ultrapure water to the ultrafiltration tube, centrifuge at 14,000 g for 10 min, and repeat 3 times. Through the above method, the protein can be desalted, and at the same time, the denaturation and modification of the sugar chain can be real...
Embodiment 3
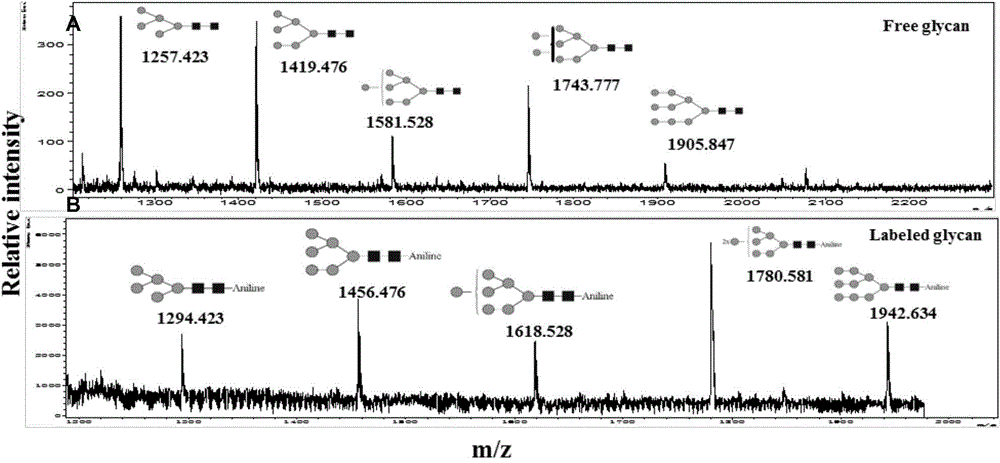
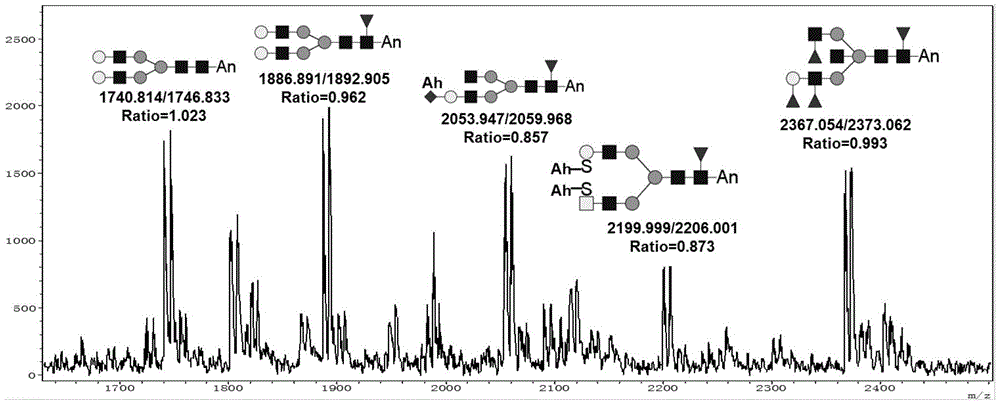
[0043] Example 3 N-linked sugar chain modification and mass spectrometry analysis of standard glycoprotein RNase B
[0044] For the modification of RNase B sugar chains, the glycoprotein RNase B is firstly modified with the amidating reagent acetylhydrazide, then the glycosidase PNGase F is used to enzymatically release the sugar chains, and then the reducing end of the N-linked sugar chains is treated with the aminating reagent aniline. Modified, the obtained sugar chain was identified by MALDI-TOF, and the [M+Na] of RNase B was obtained + sugar chain peak spectrum. At the same time, the sugar chains of RNase B released without any sugar chain modification were compared, and the results were as follows figure 2 shown, from figure 2 In -A, it can be seen that the sugar chain spectrum of RNase B is fully identified. From the spectrum, it can be seen that the sugar chains of RNase B on the same glycosylation site are slightly heterogeneous, and there are 5 high-mannose glyco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com