LAMP method for detecting bacterial resistance to florfenicol
A florfenicol and drug resistance technology, applied in the field of microbial detection, can solve the problems of losing the effect of inhibiting or killing bacteria, increasing human factors, contaminating the laboratory, etc., and achieving the effect of a simple method of determination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Establishment of a LAMP method for detecting bacterial resistance to florfenicol (floR gene)
[0028] 1. Preparation of materials
[0029] Florfenicol-resistant bacteria include Streptococcus, Escherichia coli, and Enterococcus faecalis (the MIC values for Florfenicol are 128 μg / mL, 256 μg / mL, and 512 μg / mL, respectively); Florfenicol-sensitive bacteria include Pasteurella Bacillus standard strain ATCC21955, Pasteurella, and Streptococcus (the MIC values for florfenicol are 2 μg / mL, 1 μg / mL, and 2 μg / mL, respectively) were all isolated (or purchased) and preserved by Guangxi Veterinary Research Institute. The bacterial plasmid DNA extraction kit was purchased from Beijing Kangwei Century Biotechnology Co., Ltd., and the deoxyribonucleic acid amplification kit (LAMP method, SLP206) was purchased from Beijing Lanpu Biotechnology Co., Ltd.
[0030] 2. LAMP primers
[0031] According to the bacterial florfenicol-resistant floR gene sequence in GenBank, a set...
Embodiment 2
[0052] The specificity of embodiment 2LAMP detection method
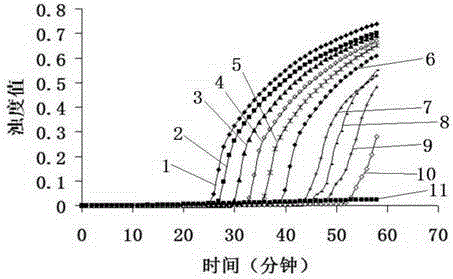
[0053] Using the recombinant plasmid pMD18-T-floR as the positive control and sterilized ultrapure water as the negative control, 3 strains of florfenicol-resistant bacteria and 3 strains of negative control bacteria were used for LAMP amplification, and the results were as follows: figure 1 As shown, the rising curve of turbidity appeared in the positive control reaction tube after 20 minutes, and the rising curve of turbidity appeared after 30 minutes in the reaction tubes of three strains of bacterial florfenicol, which was a positive result, while the three strains of negative control There was no amplification in the bacteria and water control reaction tubes, and the result of LAMP was negative.
Embodiment 3
[0054] The sensitivity of embodiment 3LAMP detection method
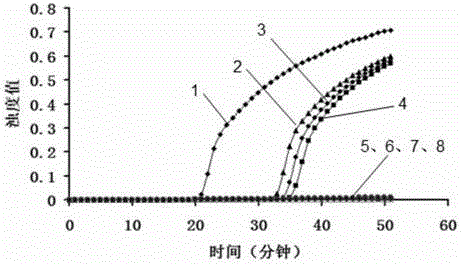
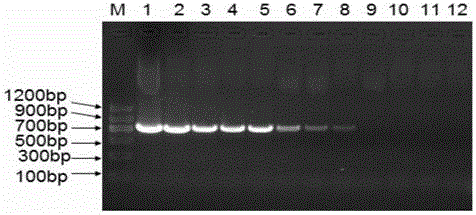
[0055] The initial concentration of the recombinant plasmid pMD18-T-floR was 2.36×10 2 ng / μL, 10-fold dilution for detection, the results are as follows figure 2 with image 3 As shown, the detection limit of LAMP method is about 2.36×10 -11 ng / μL, while the detection limit of PCR method is 2.36×10 -9 ng / μL. As the concentration of the sample decreases, the time required for the turbidity value of the LAMP amplification reaction to reach 0.1 increases.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com