Method for signal amplification and detection on target deoxyribonucleic acid (DNA) sequence at normal temperature
A technology of DNA sequence and signal amplification, which is applied in the field of single nucleotide polymorphism (SNP) typing and low-abundance point mutation detection, DNA sequence detection, and can solve problems such as complexity and insufficient discrimination
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1
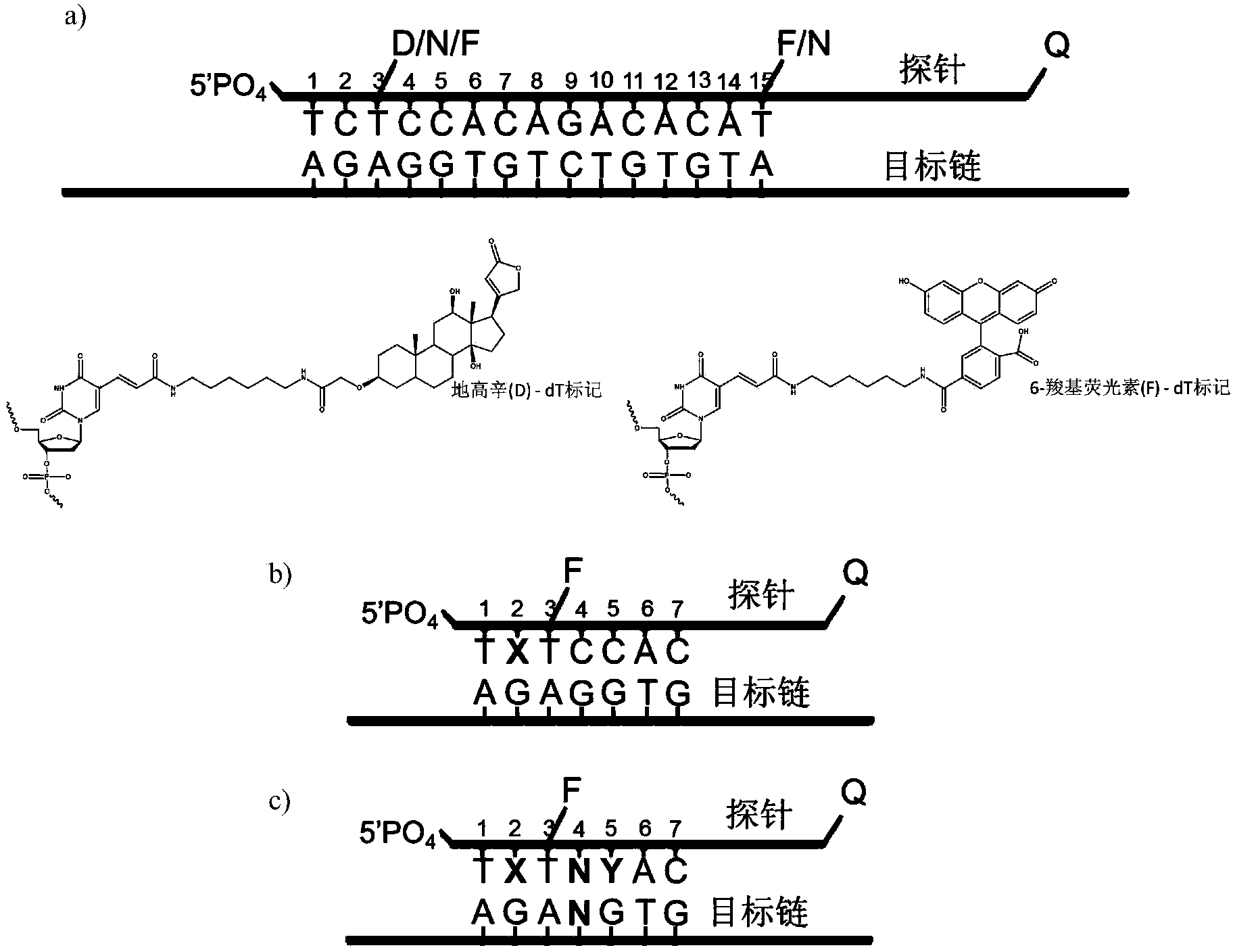
[0054] like figure 2 As shown in b), in this embodiment, the target object is a single strand of DNA. 5'-PO 4 The fluorescent group FAM in the single-stranded double-labeled fluorescent nucleic acid probe at the end is labeled on the base of the third nucleotide from the 5' end, and the quenching group BHQ1 is labeled at the 3' end. Prepare different concentrations of target DNA solutions respectively. The specific steps of detection are as follows:
[0055] In 1× ThermoPol buffer solution (20mM Tris-HCl+10mM (NH 4 ) 2 SO 4 +10mMKCl+2mM MgSO 4 , pH 8.825°C), mix the probe with different concentrations of target DNA, perform temperature-rising annealing (heat program 85°C 90s, 65°C 90s, 50°C 90s, 37°C 180s), add Lambda exonuclease, rapidly Measure the change of the fluorescence value of the solution with time.
[0056] In this embodiment, the designed probe sequence is as follows:
[0057] 5'-PO 4 -TCT(-FAM)CCACAGACACATACTCCA-BHQ1-3' (SEQ ID No. 1...
Embodiment 2
[0068] Example 2
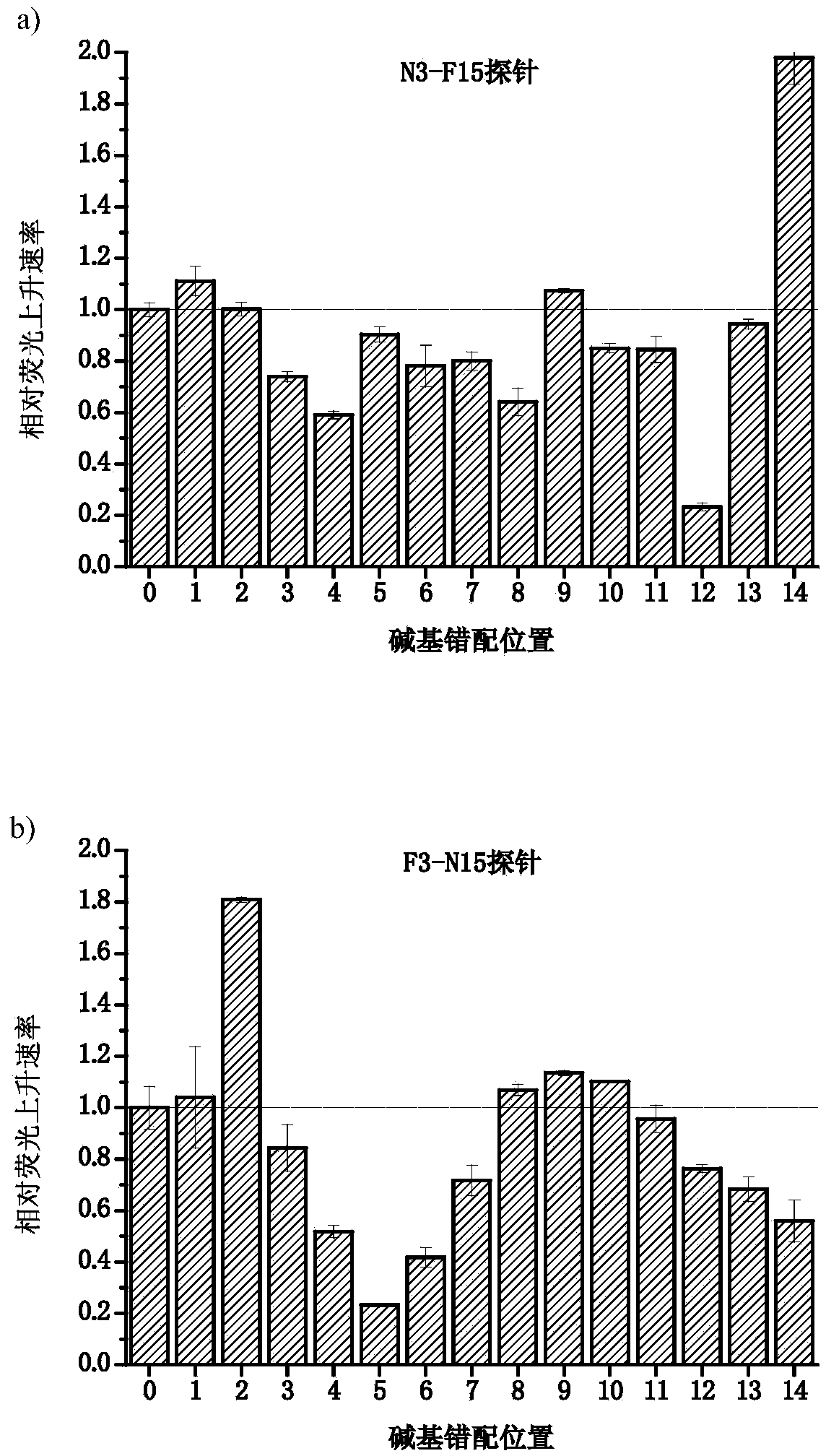
[0069] In this example, the target is a single strand of DNA. 5'-PO 4 The fluorescent group FAM in the single-stranded double-labeled fluorescent nucleic acid probe at the end is labeled on the base of the third nucleotide from the 5' end, and the quenching group BHQ1 is labeled at the 3' end. The 2nd and 5th positions from the 5' end of the probe are mismatched with the target DNA single strand, and the rest of the bases are matched. There is also an interfering strand with a single base difference from the target DNA strand in the sample. The position of the single base corresponds to the 4th nucleotide from the 5' end of the probe, so the interfering strand and the probe are at the 2nd, 4th, and 5th nucleotides. Bits are mismatched.
[0070] This example contains two experiments:
[0071] (1) Differentiation degree of different types of mismatches
[0072] The discrimination of different types of mismatches was determined by changing the base type o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com