A genetically engineered bacterium producing α-ketobutyric acid and its application
A technology of genetically engineered bacteria and ketobutyric acid, applied in the biological field, can solve problems such as complex reaction conditions, high production costs, and heavy pollution, and achieve the effects of simple fermentation process, low production cost, and high acid production rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
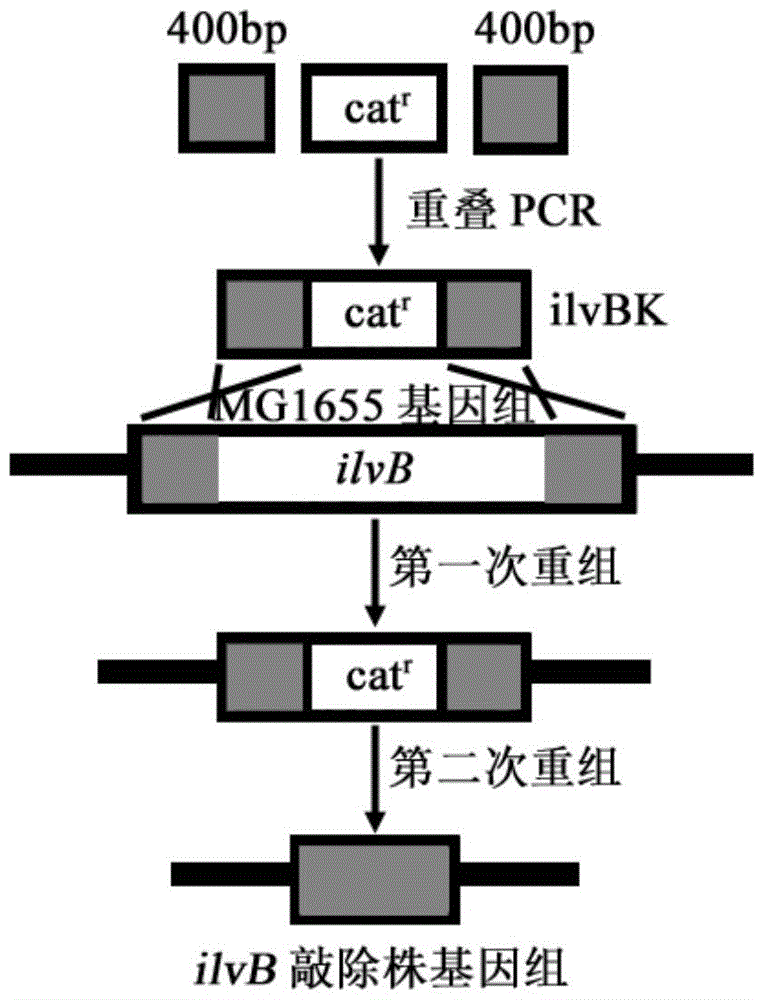
[0067] Example 1: E.coli MG1655 acylhydroxy acid synthase I large subunit coding gene ilvB knockout
[0068] According to the 5'and 3'400bp sequence of the ilvB (GeneID:948181) gene of Escherichia coli MG1655 in the NCBI database, the upper and downstream homology arm amplification primers ilvB1-F, ilvB1-R, ilvB3-F and ilvB3 were designed -R, and use the genomic DNA of the bacteria as a template to amplify homologous arm fragments.
[0069] The PCR conditions were 94°C for 5min, 1 cycle, 94°C for 30s, 55°C for 30s, 72°C for 40s, 30 cycles, 72°C for 10min, 1 cycle, and the reaction system was 100μL.
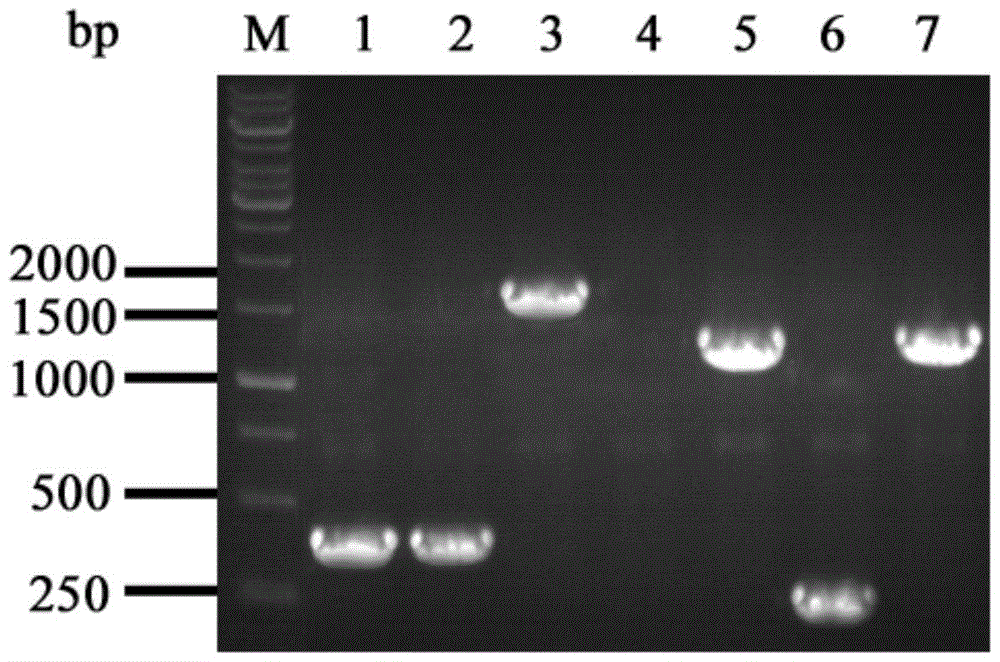
[0070] The PCR product was subjected to 1.5% agarose gel electrophoresis and then gelled and recovered, and the obtained fragments were named ilvB1 and ilvB3.
[0071] The amplification primers ilvB2-F and ilvB2-R were designed according to the chloramphenicol resistance gene cassette sequence in the plasmid pKD3, and the chloramphenicol resistance gene cassette fragment was amplified usi...
Embodiment 2
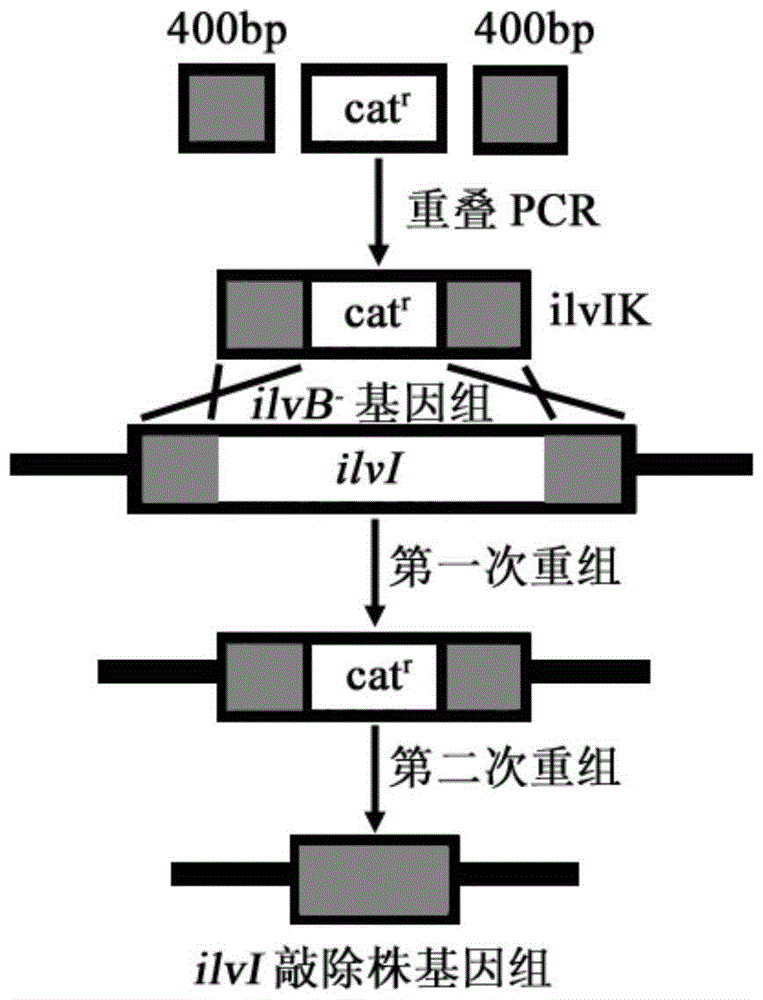
[0081] Example 2: E.coli MG1655ilvB knockout strain acetohydroxy acid synthase III large subunit coding gene ilvI knockout
[0082] Design homology arm amplification primers ilvI1-F, ilvI1-R, ilvI3-F and ilvI3-R according to the 5'and 3'400bp sequences of Escherichia coli MG1655ilvI (GeneID:947267) gene in NCBI database, and The genomic DNA of the bacteria is used as a template to amplify homologous arm fragments.
[0083] The PCR conditions were 94°C for 5 minutes, 1 cycle, 94°C for 30s, 55°C for 30s, 72°C for 40s, 30 cycles, and 72°C for 10 minutes, and the reaction system was 100 μL.
[0084] The amplification primers ilvI2-F and ilvI2-R were designed according to the sequence of the chloramphenicol resistance gene cassette in the plasmid pKD3, and the plasmid was used as a template to amplify the chloramphenicol resistance gene cassette fragment. The PCR conditions were 94°C for 5 min and 1 cycle. 30 cycles of 94°C for 30s, 55°C for 30s, 72°C for 70s, and 72°C for 10min for 1 cy...
Embodiment 3
[0090] Example 3 Construction of THRZ strain
[0091] Design homology arm amplification primers thrL1-F, thrL1-R, thrL3-F and thrL3-R according to the 5'and 3'end 400bp sequences of thrL (GeneID:948283) gene in E.coli MG1655 in NCBI database, and The genomic DNA of the bacteria was used as a template to amplify homologous arm fragments.
[0092] The PCR conditions were 94°C for 5 minutes, 1 cycle, 94°C for 30s, 55°C for 30s, 72°C for 40s, 30 cycles, and 72°C for 10 minutes, and the reaction system was 100 μL.
[0093] The PCR products were subjected to 1.5% agarose gel electrophoresis and then gelled and recovered. The obtained fragments were named thrL1 and thrL3.
[0094] The amplification primers thrL2-F and thrL2-R were designed according to the chloramphenicol resistance gene cassette sequence in the plasmid pKD3, and the chloramphenicol resistance gene cassette fragment was amplified using the plasmid as a template.
[0095] The PCR conditions were 94°C for 5 minutes, 1 cycle, 94...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com