Method for changing phosphorylation site of rice phosphate transporter gene OsPT8 and application thereof
A technology of transporter and phosphate, applied in the field of genetic engineering and crop genetic improvement, can solve the problems of hindering the normal output of PHT1 protein and affecting the effective accumulation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Embodiment 1, OsPT8 site-directed mutation
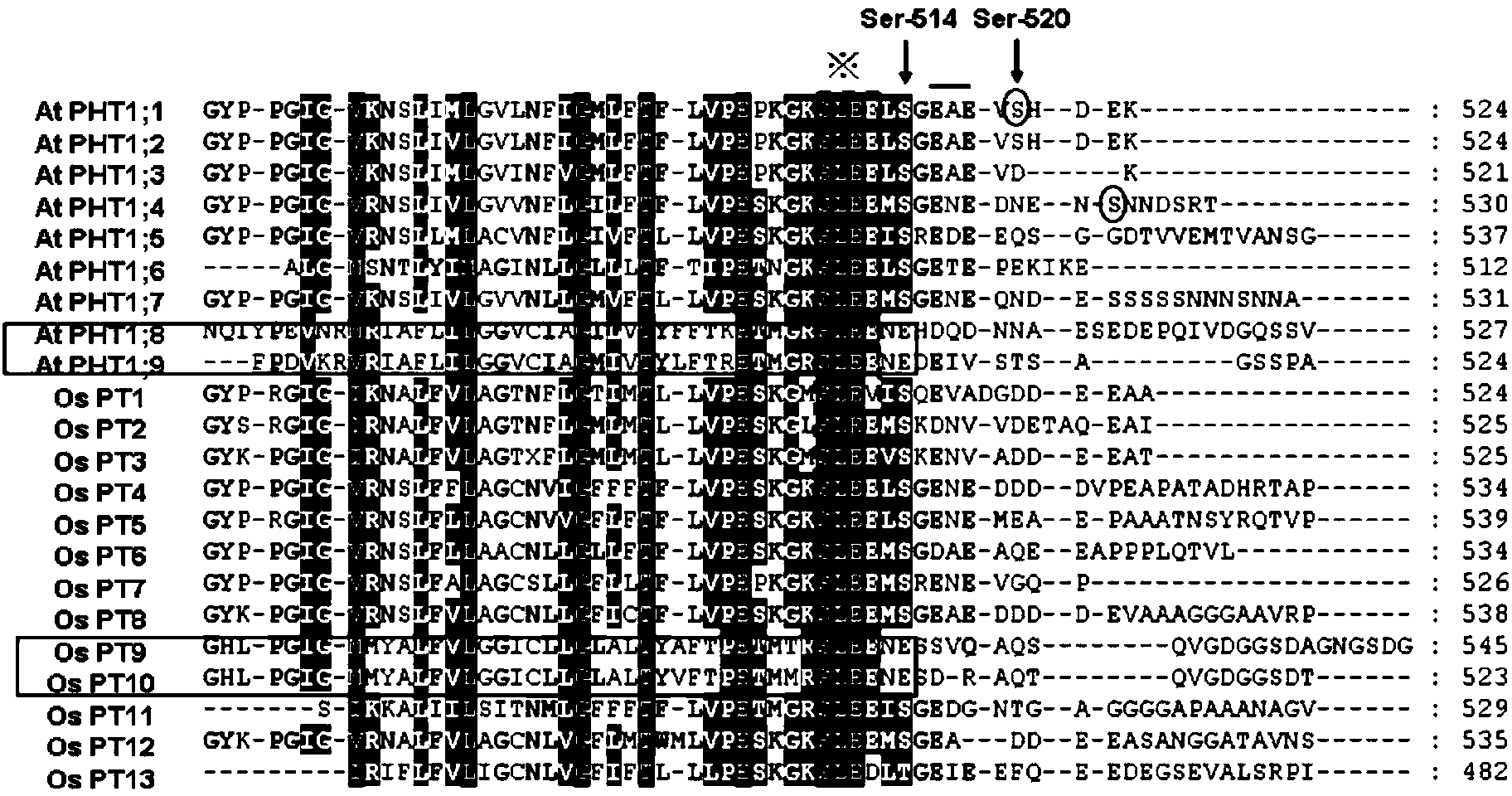
[0046] 1. Search for possible phosphorylation sites: Download 9 PHT family members of Arabidopsis and 13 homologous proteins of rice from NCBI for sequence alignment ( figure 1 ), it was found that the 517th and 512th serines of OsPT8 (SEQ ID NO: 1) amino acids are the most likely homologous sites to the 514th serines of Arabidopsis PHT1;1.
[0047] 2. Primer design for site-directed mutagenesis: Due to the different codon preferences of proteins of different species, the Editseq software of DNASTAR Company was used to count the codons encoding alanine in OsPT8, and found that GCG in OsPT8 was the main alanine codon. Mutation of PT8Ser-517 to GCG(Ala) was therefore chosen. By designing primers with mutated bases, the primer sequences are as follows:
[0048] Table 1. Primers for point mutation of OsPT8 protein Ser
[0049] Primer name
Primer sequence (5'-3')
PT8A-P1
GCTCTAGAGCGCGGCAGGAGCAGCAGCAGCA ...
Embodiment 2
[0080] Embodiment 2, vector construction and transgene
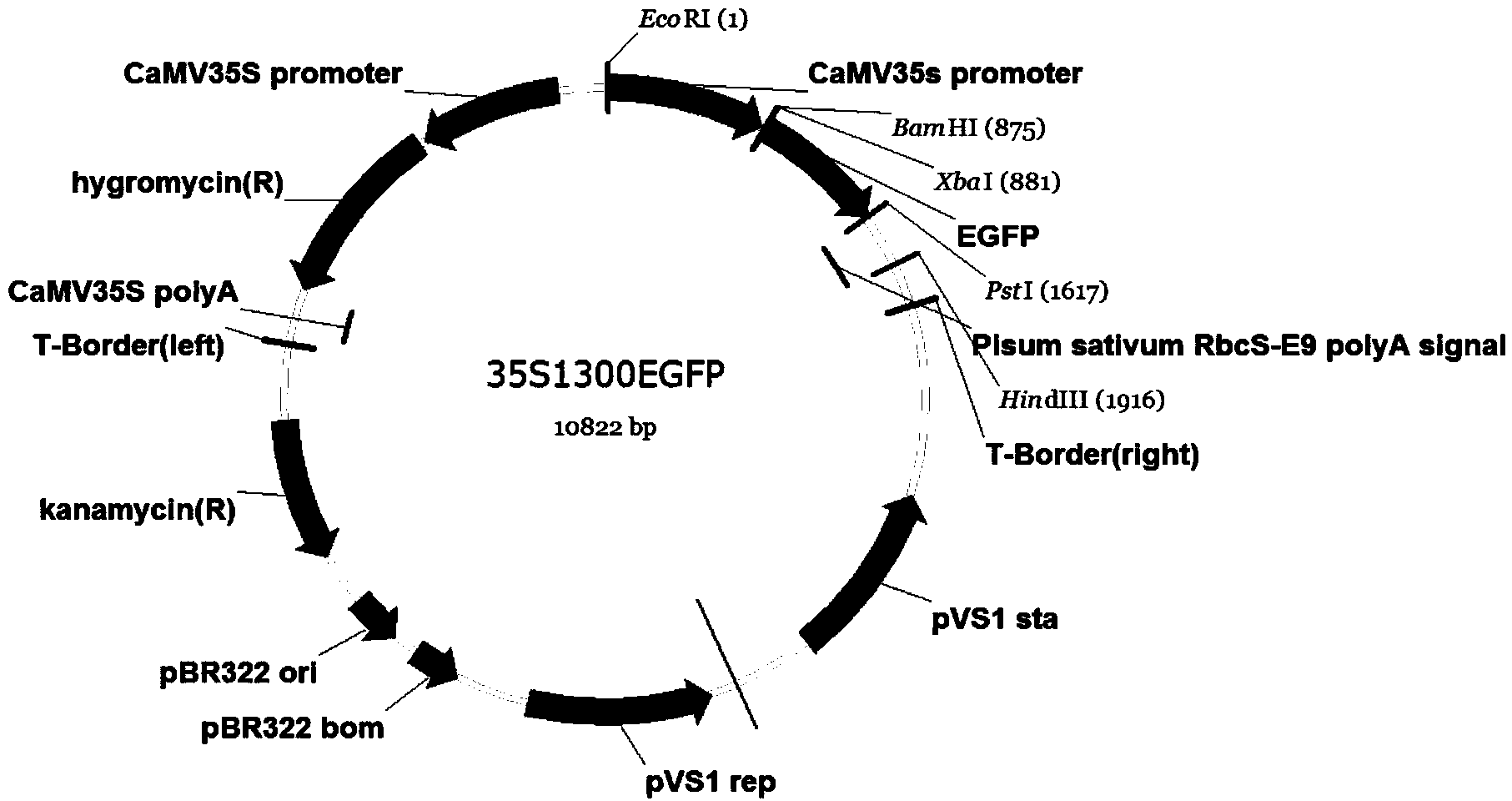
[0081] 1. Construction of the binary vector: Design primers PT8A-CDS-SalI-low and PT8A-CDS-SalI-up (Table 2) according to the CDS sequence of the OsPT8 gene, and introduce EcoR I and Sal I restriction sites (underlined ), using pCAMBIA1300-35S-OsPT8 in Example 1 S517A -GFP plasmid was used as a template to amplify OsPT8 S517A gene sequence.
[0082] The PCR reaction system is 50μl, including 100ng of DNA template, 1μl of each 10mM primer pair, 5μl of 10×PCR buffer, 5μl of 2mM dNTP mixture, 25mM MgCl 2 4 μl, add ddH 2 O to a total volume of 50 μl. The PCR amplification reaction program is: 1cycle (94°C, 10min), 29cycles (94°C, 1min; 58°C, 1min; 72°C, 1.5min), 1cycle (72°C, 5min). Amplified products were detected on 1.0% agarose gel.
[0083] Then use EcoR I and Sal I double enzyme digestion and connect into pCAMBIA1301-35S (see above for the construction method) to obtain 35S-1300-OsPT8 S517A . Use the OsPT8 promo...
Embodiment 3
[0090] Embodiment 3, available phosphorus analysis of transgenic positive strains
[0091] Take NIP wild type and transgenic positive line PT8 S517A 40 rice seeds of No. 1-3 strains were soaked at 37°C until they were white. The seeds of Lubai were sown in the nutrient solution with 100 M phosphorus concentration, and grown in the growth room (temperature 30°C during the day, 22°C at night, light 350mmol m- 2 S -1 ) After 7 days, five plants were transplanted into 10L of 100M phosphorus concentration nutrient solution (see Table 3 for formula), and continued to grow in the growth room (temperature 30°C during the day, 22°C at night, light 350mmol m- 2 the s -1 ) for 21 days, and the nutrient solution was replaced once in the middle. The other positive seedlings were harvested in the greenhouse and used to screen pure strains. After observing the phenotype, the shoots and roots were taken respectively (the roots were rinsed with deionized water, and finally blotted dry wit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com