Saccharomyces cerevisiae engineering bacteria with low-yielding ethyl carbamate, and building method and application of saccharomyces cerevisiae engineering bacteria
A technology of urethane and Saccharomyces cerevisiae, applied in the field of yeast metabolic engineering bacteria and its construction, to achieve the effect of reducing the formation of EC and improving the utilization capacity of urea
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
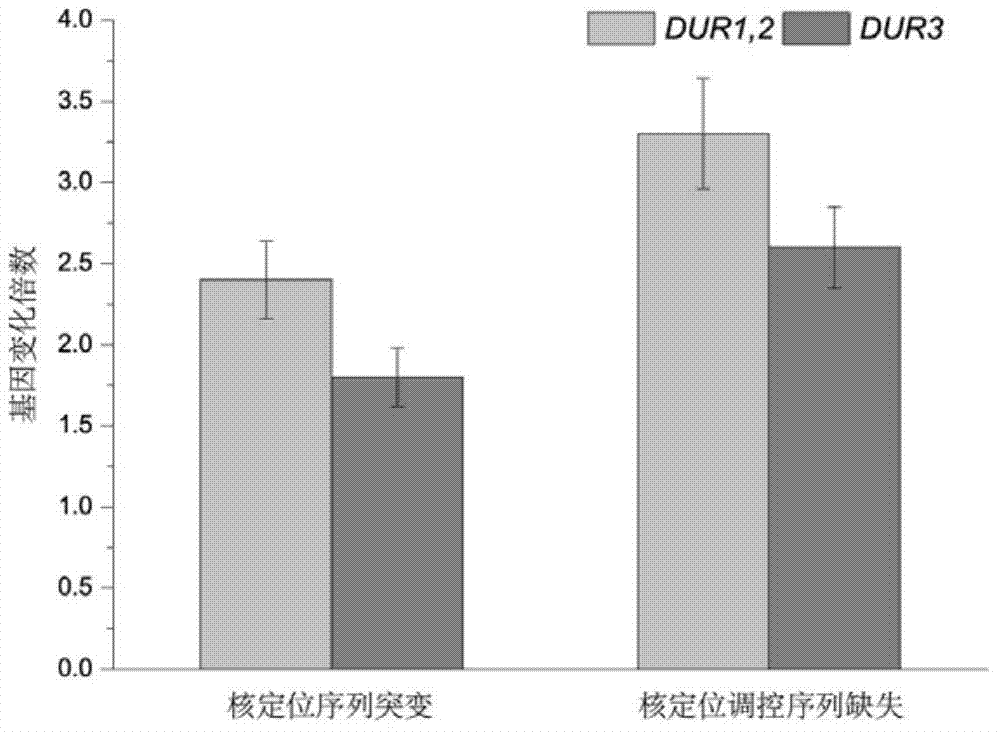
[0021] Example 1 The effect of the mutation of the regulatory factor Gln3p nuclear localization sequence on the expression of urea metabolism genes in bacterial strains
[0022] The build method is as follows:
[0023] 1) A GLN3 gene fragment ( figure 1 ). The GLN3 gene amplified by PCR was recovered and purified by gel, ligated with pMD19-T Vector (Takara), and transformed into E.coli JM109. The transformed positive clone T-GLN3 was selected by colony PCR on the ampicillin-resistant plate and sent to Shanghai Sangon for sequencing verification. Store the strains with correct sequences verified by sequencing in glycerol tubes at -20°C;
[0024] 2) After activating the strain saved in the previous step, extract the T-GLN3 plasmid. Using the T-GLN3 plasmid as a template and the mutation primers in Table 1 as primers, according to the instructions of the MutanBEST point mutation kit of Bao Bio Company, mutate the 344th, 347th and 355th Ser sites on Gln3p to alanine acid. Th...
Embodiment 2
[0027] Example 2 The effect of deletion of regulatory factor Gln3p nuclear localization regulatory sequence on strain urea metabolism
[0028] The build method is as follows:
[0029] 1) GLN3 with a length of 1959bp was amplified from the genome of Saccharomyces cerevisiae using GLN3-F' and GLN3-R' (see Table 1 for details) primers 1-653 Gene fragment( figure 1 ). GLN3 obtained by PCR amplification 1-653 After the gene was recovered and purified by gel, it was ligated with pMD19-TVector (Takara) and transformed into E.coli JM109. The transformed positive clone T-GLN3 was selected by colony PCR on the ampicillin-resistant plate 1-653 , sent to Shanghai Sangon for sequencing verification. Store the strains with correct sequences verified by sequencing in glycerol tubes at -20°C;
[0030] 2) Extract the constructed T-GLN3 1-653 And the preserved pYX212 plasmid, after Nco I and Sac I double digestion, gel recovery respectively. The recovered GLN3 1-653 The fragment was li...
Embodiment 3
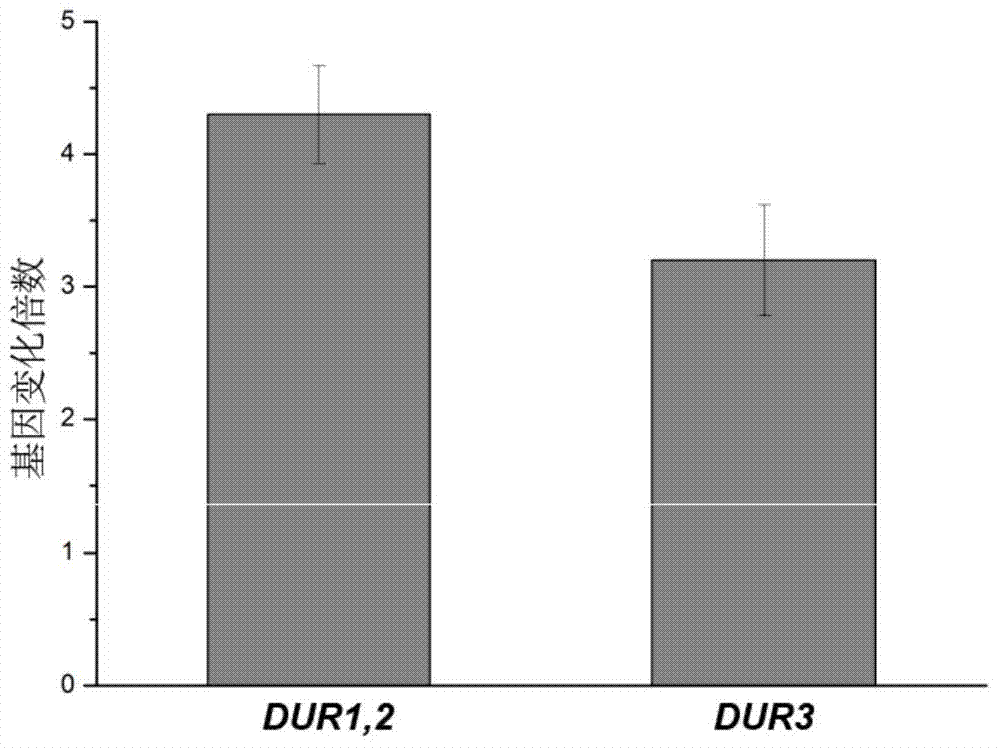
[0032] Embodiment 3 Construction of more optimal low-yield EC yeast engineering bacteria
[0033] On the basis of the positive results of both the mutation of the Gln3p nuclear localization sequence and the nuclear localization regulatory sequence, the two methods were combined to construct a better yeast engineering strain with low EC production. The build method is as follows:
[0034] With the T-GLN3 constructed in embodiment 2 1-653 The plasmid was used as a template, and the mutation primers in Table 1 were used as primers. The Gln3p 1-653 Ser sites 344, 347 and 355 were mutated to alanine. The resulting T-GLN3 1-653,S344A,S347A,S355A The plasmid was transformed into E.coli JM109 and sent to Shanghai Sangon for sequencing verification. The strains with the correct sequence verified by sequencing were stored in glycerol tubes at -20°C.
[0035] In order to test the effect of deletion of the Gln3p nuclear localization regulatory sequence, real-time quantitative PCR was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com