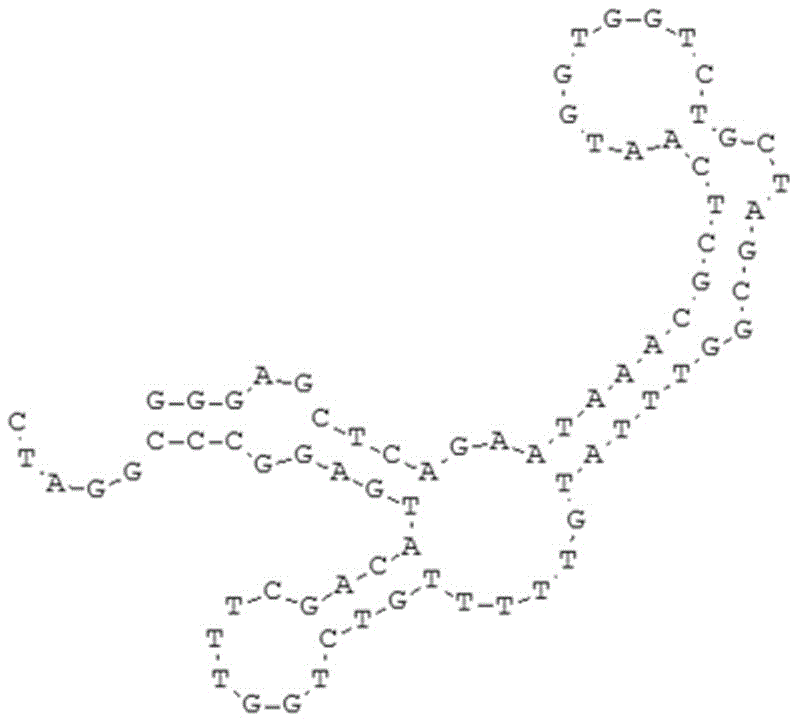
Aeromonas hydrophila aptamer and its screening method and application
A technology of Aeromonas hydrophila and aptamer, which is applied in the field of Aeromonas hydrophila aptamer and its screening, which can solve the problems of cumbersome detection process, high cost, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
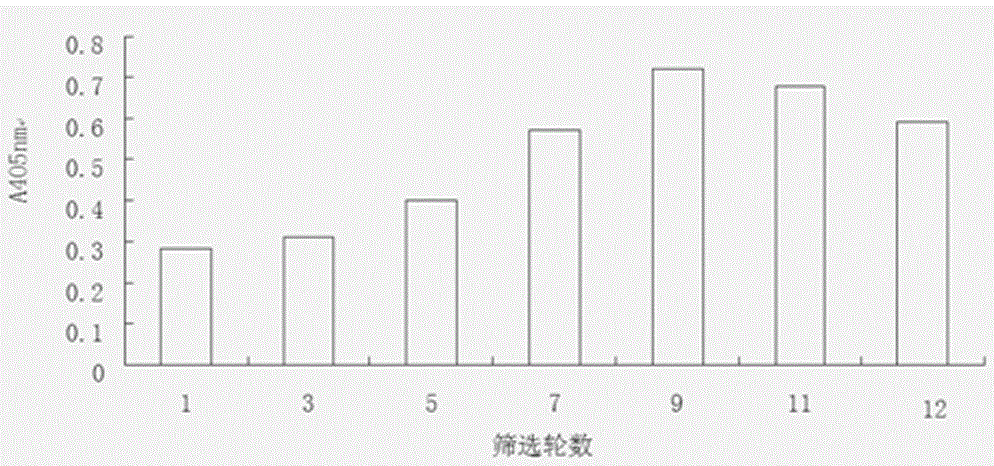
Method used
Image
Examples
Embodiment Construction
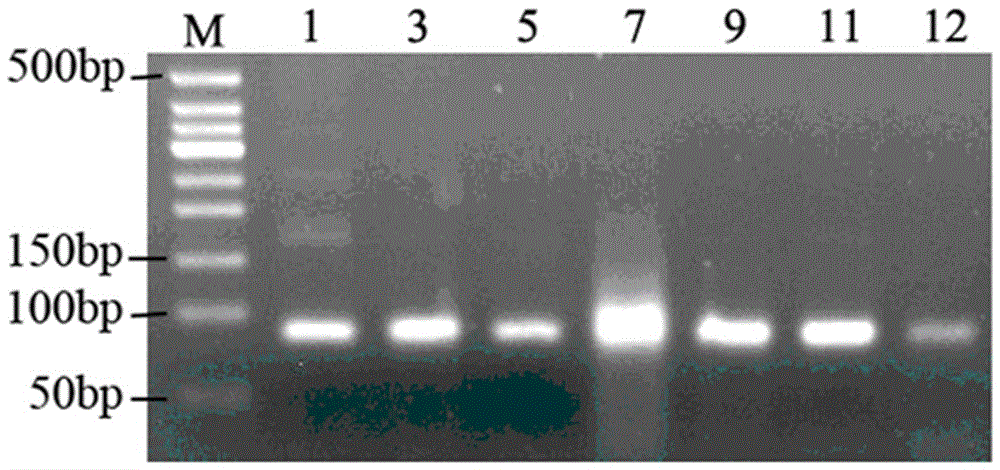
[0023] 1. Synthesis of ssDNA library and primers
[0024] Referring to the method disclosed in the literature (7. Liu Fengwei, Lan Xiaopeng. In vitro screening of Pseudomonas aeruginosa aptamer research and preliminary application [D]. Fuzhou Fujian Medical University, 2007.), the random ssDNA library used for SELEX screening and primers were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0025] The library used was: 5'-GGGAGCTCAGAATAAACGCTCAA-N35-TTCGACATGAGGCCCGGATC-3'.
[0026] The primers used are:
[0027] Primer Ⅰ: 5'-GGGAGCTCAGAATAAACGCTCAA-3';
[0028] Primer II: 5'-GATCCGGGCCTCATGTCGAA-3';
[0029] Primer III: 5′-Digoxigenin-GGGAGCTCAGAATAAACGCTCAA-3′;
[0030] Primer IV: 5'-Biotin-GATCCGGGCCTCATGTCGAA-3'.
[0031] After the above primers were synthesized, they were diluted with TE buffer (pH 8.0) commonly used in molecular biology to a concentration of 10 μmol, and stored at -20°C for future use.
[0032] 2. Experimental reagents a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com