Method for treating textile with endoglucanase
An endoglucanase and textile technology, applied in the field of textile manufacturing, can solve the problems of fading, discoloration of denim products, etc., and achieve the effects of increasing abrasion, lowering the level of color reversion and saving energy.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0204] Example 1: Cloning of Tt Cel45a(CBM+) and Tt Cel45a(CBM-) from Genomic DNA
[0205] The wild-type GH45 endoglucanase gene was cloned from the genomic DNA of Thielavia terrestris NRRL8126 as described in Example 1A of WO96 / 29397 (incorporated herein by reference).
[0206] Thielavia terrestris was grown on PDA agar plates at 37°C for 4 to 5 days. The mycelium was collected directly from the agar plate into a sterilized mortar and frozen under liquid nitrogen. Grind the frozen mycelium into a fine powder with a mortar and pestle, and use Genomic DNA was isolated using the Plant Mini Kit (QIAGEN Inc., Valencia, CA, USA).
[0207] Mutations were then performed according to Example 1 of WO98 / 12307 (incorporated herein by reference), wherein glutamine at position 119 ( Q) is substituted with histidine (H). This mutation corresponds to Q141H in SEQ ID NO: 2 of the present invention. The PCR fragment was ligated into pGEM-T (Promega Corporation, Madison, WI, USA). In thi...
Embodiment 2
[0222] Embodiment 2: the expression of Tt Cel45a (CBM+) and Tt Cel45a (CBM-) gene in Aspergillus oryzae
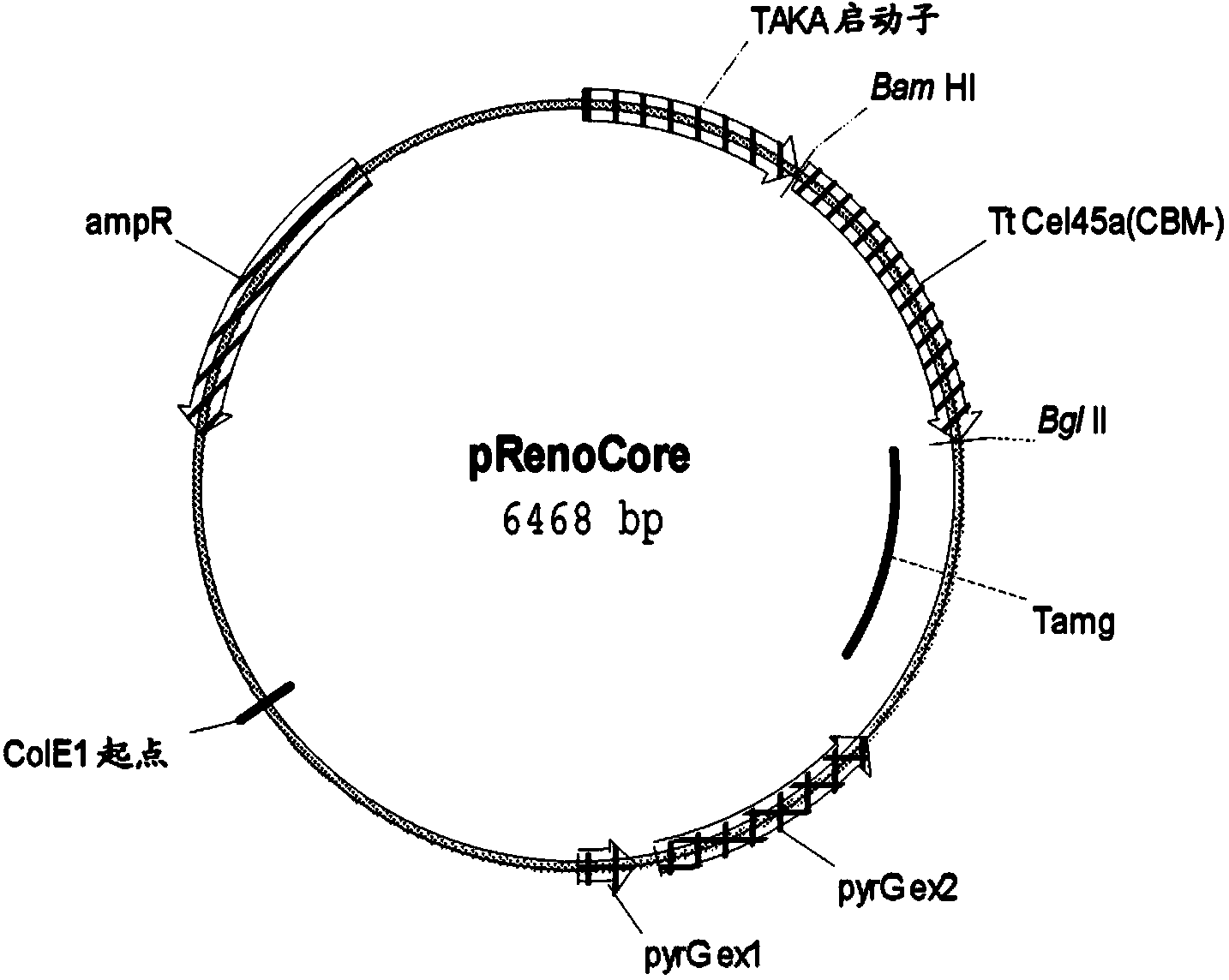
[0223] Aspergillus oryzae HowB101 (described in WO9535385 Example 1, incorporated herein by reference) protoplasts were prepared according to the method described in Christensen et al., 1988, Bio / Technology 6:1419-1422. Aspergillus oryzae HowB101 was transformed with 3 μg of pRenoCBD or pRenoCore.
[0224] Transformation of Aspergillus oryzae HowB101 with pRenoCBD or pRenoCore each produced approximately 50 transformants. The four transformants were segregated to separate minimal medium plates.
[0225] The four transformants were respectively inoculated into 3ml of YPM medium (1% yeast extract, 2% peptone and 2% maltose) in a 24-well plate, and incubated at 30°C, 150rpm. After 3 days of incubation, 20 μl of supernatant from each culture was analyzed on NuPAGE Novex 4-12% Bis-Tris Gel w / MES (Invitrogen Corporation, Carlsbad, CA, USA) according to the manufacturer's instr...
Embodiment 3
[0227] Example 3: Purification of mature polypeptides of Tt Cel45a (CBM+) and Tt Cel45a (CBM-)
[0228] 3000 ml supernatant of transformant-2 of strain-1 described in Example 2 was precipitated with ammonium sulfate (80% saturated) and redissolved in 100 ml of 25 mM Tris-HCl buffer, pH 7.0, and then against the same buffer Dialyzed and filtered through a 0.45 nm filter to a final volume of 200 ml. The solution was loaded onto a 50 ml Q FF column equilibrated in 25 mM Tris-HCl buffer, pH 7.0, and the protein was eluted with a linear NaCl gradient (0 - 0.4 M). Fractions with activity against AZCL-β-glucan (available from Megazyme for a substrate for endoglucanases) were pooled. The pooled solutions were then concentrated by ultrafiltration. The purified mature polypeptide of Tt Cel45a (CBM+) was at least 95% pure as judged by SDS-PAGE analysis.
[0229] 3000 ml supernatant of transformant-2 of strain-2 described in Example 2 was precipitated with ammonium sulfate (80% saturat...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com