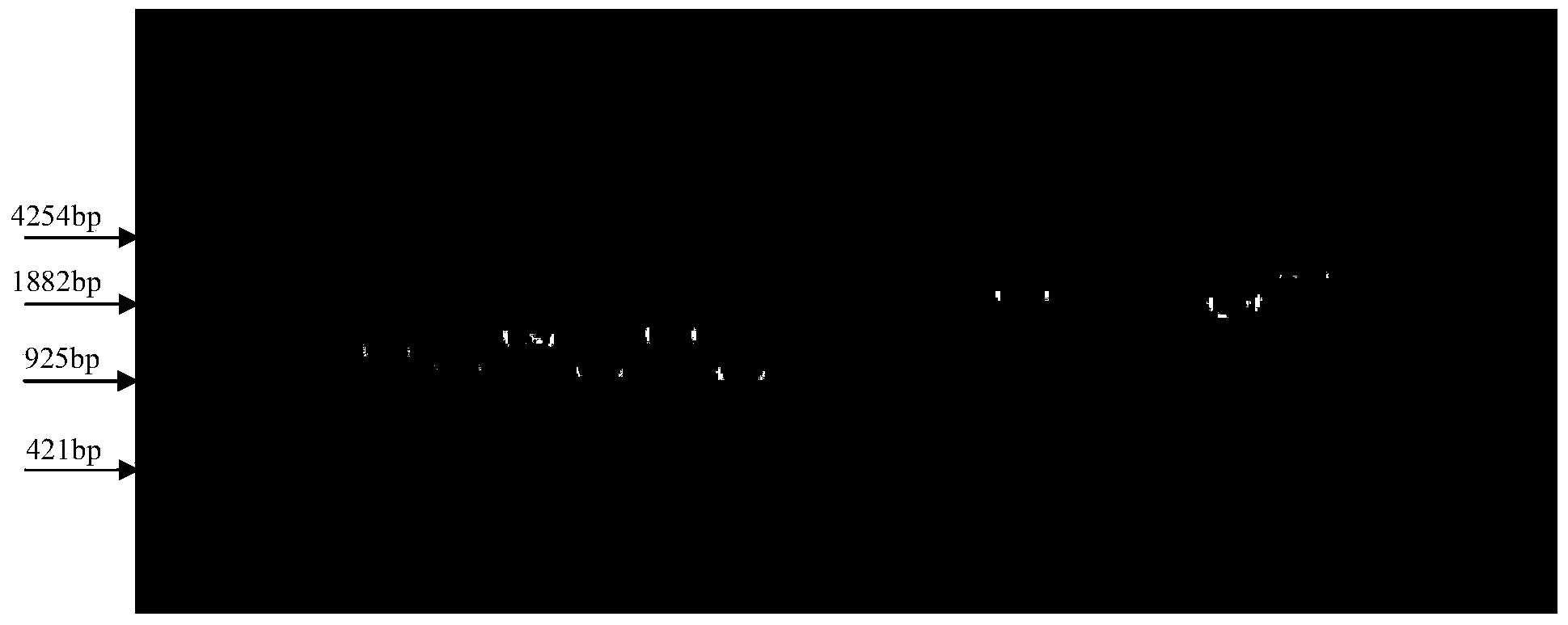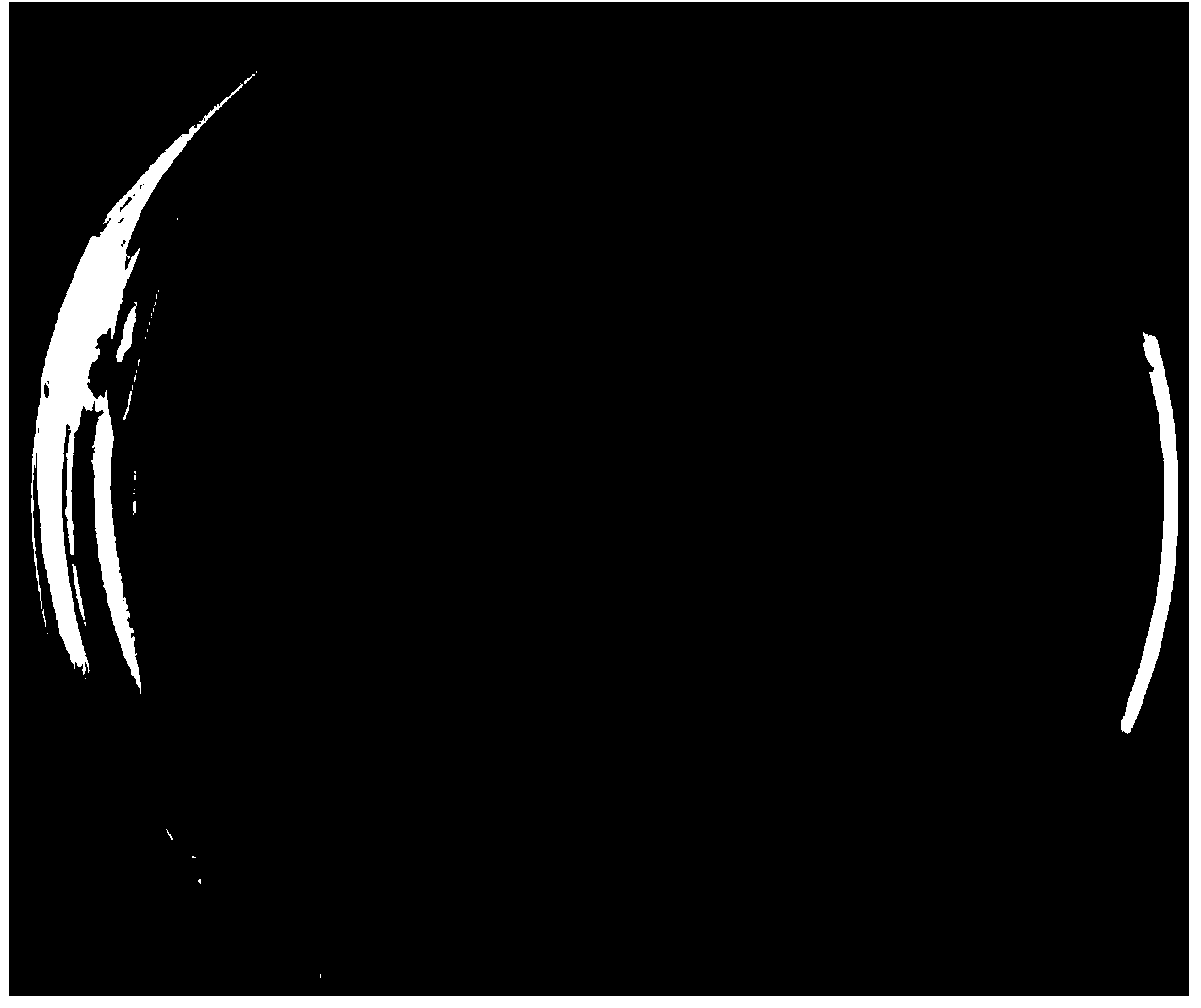Phosphate solubilizing gene for promoting acetic acid secretion
A phosphorus-dissolving gene and gene technology, applied in the field of phosphorus-dissolving genes that promote the secretion of acetic acid, can solve the problem of no functional analysis and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The cDNA full-length library construction of embodiment 1 Aspergillus niger
[0034] The strain Aspergillus niger strain H1 used in the experiment was screened from soil samples by the Agricultural Strain Collection Center of the Institute of Agricultural Resources and Agricultural Regional Planning, Chinese Academy of Agricultural Sciences. It was cultured on the insoluble phosphorus inorganic salt medium for 3 days, and about 0.2 g of fresh mycelia (such as figure 1 ), ground with liquid nitrogen, extracted total RNA with RNAisoplus kit, took 1 μg of RNA as a template for synthesizing the first strand of cDNA, guided by 3'SMART CDS Primer IIA and SMARTer IIA oligonucleotide primers, passed SMARTscriptTM reverse transcriptase The first-strand cDNA was synthesized by reverse transcription at 70°C for 2 min; double-stranded cDNA was synthesized by LD-PCR using 2 μL of the first-strand cDNA product as a template. Use CHROMA SPIN+TE-1000 to purify cDNA, remove fragments sm...
Embodiment 2
[0035] Example 2 Screening and Bioinformatics Analysis of Phosphorus-Solubilizing Genes
[0036] Dilute the culture solution containing the bacteria in the library onto the insoluble phosphate medium containing ampicillin (100 μg / mL), incubate at 37°C for 3-4 days, observe whether there is a phosphate-dissolving transparent circle, and transfer the transformant that produces the transparent circle to cultivated, such as image 3 As shown, the screened clones can still produce transparent circles on the insoluble phosphorus inorganic medium after transfer, indicating that their functions are stable. The stable colonies were extracted from the plasmids and sequenced to obtain DNA sequences, which were compared by Blast on NCBI Analysis, and use DNAMAN6.0 to analyze the open reading frame, and compare and analyze the translated protein on the PDB database.
Embodiment 3
[0037] Construction of embodiment 3 expression vector, transformation host, positive clone identification
[0038] Primer design (Forward: 5'-TATTCGGAATTCATGTCTGCCAATCGAGCAAG-3'; Reverse: 5'-CAAGTACTCGAGCTACGGCTCGCCGAGGAGCCG-3'), using high-fidelity Taq enzyme to amplify the ORF of the above gene, reaction conditions: 94°C for 5min; 94°C for 1min, 60°C for 1min , 72°C 1min cycle 35 times 72°C 10min, 4°C forever. After double digestion with restriction endonucleases EcoRI and XhoI, it was connected to the expression vector pGEX-6p. The sequence of the restriction site and the map of the expression vector pGEX-6p are as follows: Figure 4 , Figure 5As shown, E.coli HST08 was transformed by electric shock method, and the clones that could produce transparent circles were screened on the insoluble phosphorus inorganic salt medium ( Figure 6 ), and the fragment size of the positively transformed clones was selected by PCR identification, and the amplification results were as fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com