shrna (short hairpin ribonucleic acid) for inhibiting expression of human oral cancer cell PRPS2 (phosphoribosyl pyrophosphate synthetase subunit ii) as well as construction and application of carrier of shrna
A technology for cancer cells and human oral cavity, which is applied in the fields of molecular biology and biomedicine to achieve the effect of inhibiting proliferation and overcoming the short duration of action.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Design of the hTERT promoter sequence of the present invention
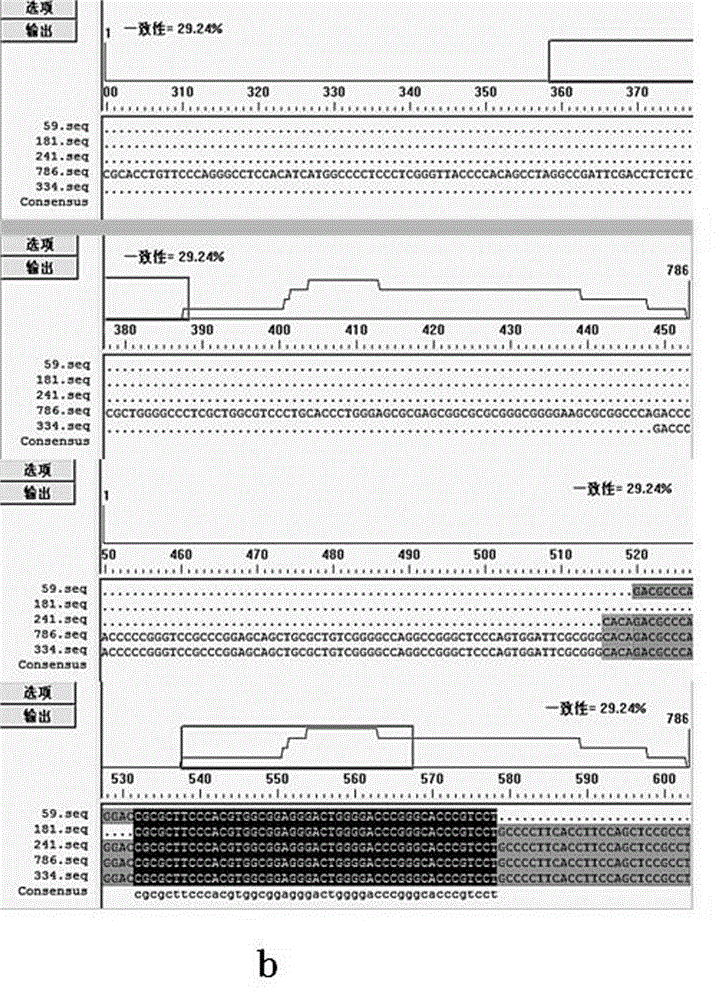
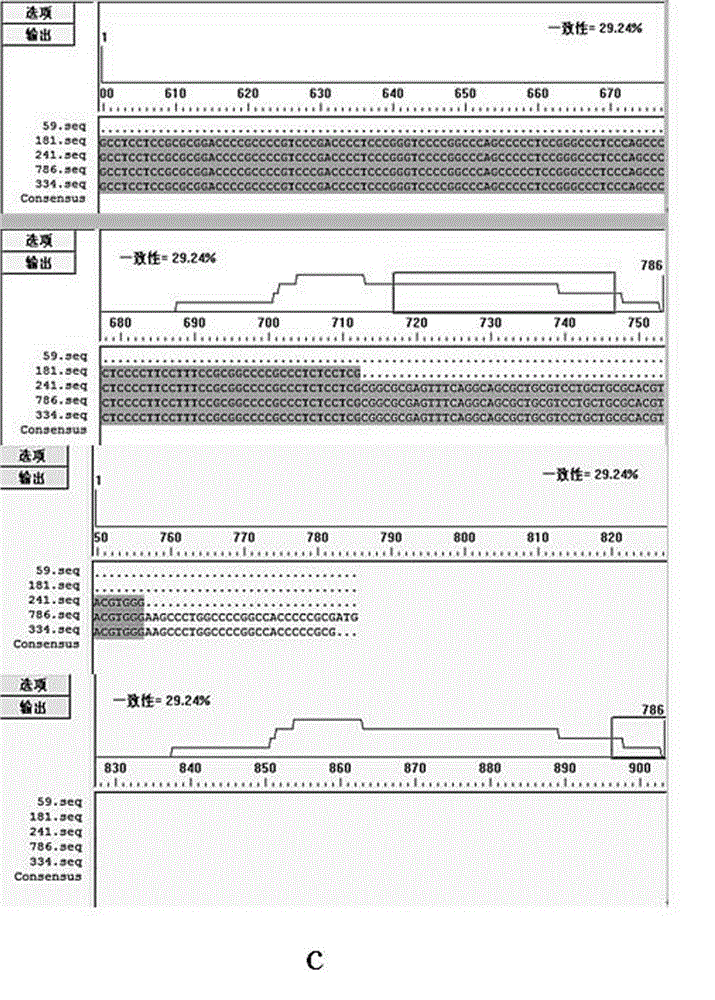
[0032] The promoter region is the binding region of RNA polymerase, its sequence directly determines the structure, and the structure is directly related to the efficiency of transcription. Using DNAMAN analysis software, the 334bp hTERT promoter sequence (SEQ ID NO: 4) designed and used in the present invention was compared with the 4 hTERT promoter sequences reported in the literature, respectively [Horikawa I, Cable PL, Afshari C, Barrett JC. Cloning and characterization of the promoter region of human telomerase reverse transcriptase gene. Cancer Res. 1999 Feb 15;59(4):826-830.] The 59bp hTERT promoter sequence reported (SEQ ID NO: 5), Literature [Takakura M, Kyo S, Kanaya T, Hirano H, Takeda J, Yutsudo M, Inoue M. Cloning of human telomerase catalytic subunit (hTERT) gene promoter and identification of proximal core promoter sequences essential for transcriptional activation in immortalized and cancer...
Embodiment 2
[0034] Design of shRNA that can specifically reduce PRPS2 gene expression in human oral cancer cells
[0035] Search the mRNA base sequence of the human PRPS2 gene in the NCBI database (GeneBank number: NM_001039091.2), and use the siRNA finder software (http: / / www.ambion.com / techlib / misc / siRNA_finder.html) to select the target Sequence, the target sequence is the 422nd to 440th positions of human PRPS2 mRNA, as shown in SEQ ID NO: 2, 5'- AUACGCCCGA CAAGAUAAA -3'. The DNA sequence that can be transcribed to generate shRNA designed according to the target sequence is: SEQ ID NO: 3: 5'- CCATACGCCC GACAAGATAA ACTCGAGTTT ATCTTGTCGG GCGTATGG -3', and the shRNA sequence generated after transcription is: SEQ ID NO: 1: 5' - CCAUACGCCC GACAAGAUAA ACUCGAGUUU AUCUUGUCGG GCGUAUGG -3'.
Embodiment 3
[0037] Construction of recombinant vector pGGN-hTERT-PRPS2 that can specifically reduce the expression of PRPS2 gene in human oral cancer cells
[0038] (1) The design scheme for recombining SEQ ID NO: 3 into pGPU6 / GFP / Neo siRNA eukaryotic expression vector is: EcoRI- Target sense sequence - Hairpin loop - Target antisense sequence - Terminator - BamHI, the specific operation steps are as follows:
[0039] Operation Step 1: Use a DNA synthesizer to synthesize the DNA shown in SEQ ID NO: 3, and add EcoRI and BamHI restriction sites at both ends. The structural model diagram is shown in figure 2 .
[0040] The second step of operation: annealing the above-mentioned artificially synthesized single-stranded DNA sequence to form a double-stranded structure. The annealing reaction system is as follows: each 2mL of forward and reverse DNA oligonucleotides is mixed with 46ml of annealing buffer, at 95°C for 4min, at 70°C for 10min, and slowly cool the annealed oligonucleotides t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com