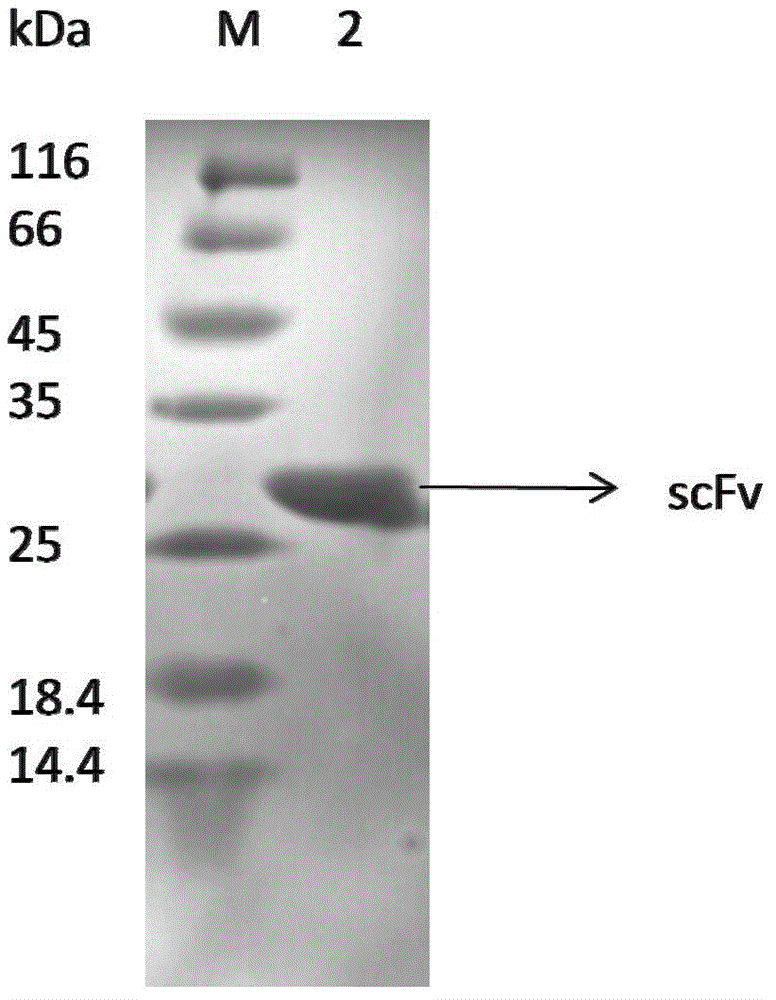
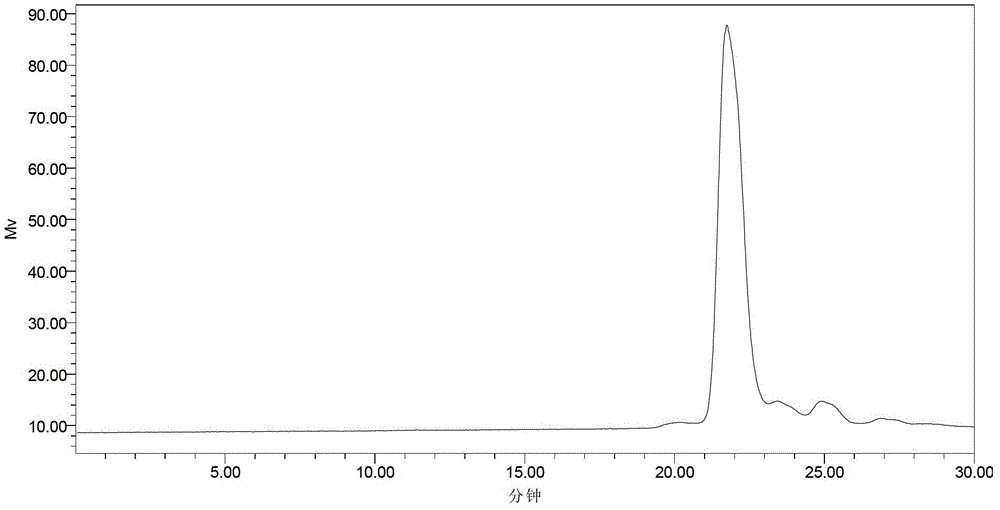
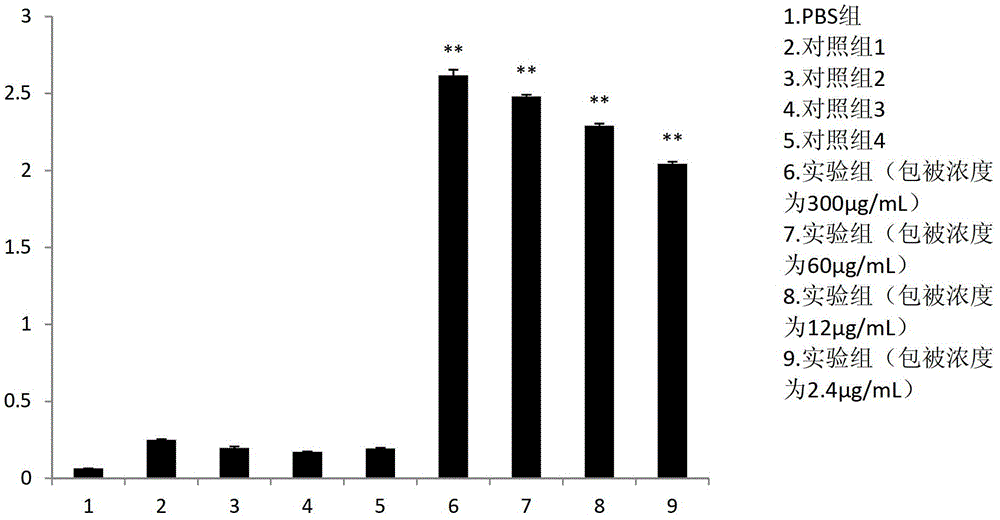
scfv antibody, encoding gene thereof and application thereof to preparation of preparation for treating or preventing infectious bursal disease
A gene and coding technology, applied in the field of scFv antibodies, can solve the problems of limitation and poor controllability of industrial production, and achieve the effect of strong specificity, avoiding horizontal transmission of diseases, and good therapeutic effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1. Discovery of scFv antibody (single-chain antibody) and its coding gene
[0031] 1. Construction of antibody library
[0032] The spleens of chickens immunized with IBDV were taken, RNA was extracted and reverse transcribed into cDNA. According to the antibody sequence on GenBank, gene primers for cloning the variable region of the antibody were designed, and cDNA was used as a template to clone the variable region of the antibody by PCR. The VH and VL fragments were respectively inserted into the upstream and downstream of the Linker of the pTlinker vector to construct a scFv antibody library. The scFv antibody library was digested and connected to the bacterial display vector pBSD to construct a bacterial surface display library of anti-IBDV antibodies. VH is about 380bp, VL is about 320bp, and VH-Tlinker-VL is about 740bp.
[0033] 2. Antibody library screening
[0034] All clones after transformation were collected, induced by IPTG (0.25mmol / L) for 4h,...
Embodiment 2
[0037] Embodiment 2, preparation of scFv antibody
[0038] 1. Synthesize the double-stranded DNA molecule shown in sequence 2 of the sequence listing.
[0039] 2. Using the double-stranded DNA molecule synthesized in step 1 as a template, perform PCR amplification with a primer pair composed of F1 and R1 to obtain a PCR amplification product.
[0040] F1: 5'–CATG CCATGG GCCGTGACGTTGGACGAG-3';
[0041] R1: 5'–CCC AAGCTT TTAACCTAGGACGGTCAGGG-3'.
[0042] 3. Digest the PCR amplified product of step 2 with restriction endonucleases NcoI and HindIII, and recover the digested product.
[0043] 4. Digest the pET-27b(+) vector with restriction endonucleases NcoI and HindIII to recover a vector backbone of about 5367 bp.
[0044] 5. Ligate the digested product of step 3 with the vector backbone of step 4 to obtain a recombinant plasmid. According to the sequencing results, the structure of the recombinant plasmid is described as follows: a double-stranded DNA molecule shown in ...
Embodiment 3
[0053] Embodiment 3, the preparation of VP2 protein
[0054] 1. Construction of recombinant plasmids
[0055] 1. Synthesize the double-stranded DNA molecule shown in sequence 4 of the sequence listing.
[0056] 2. Using the double-stranded DNA molecule synthesized in the step as a template, perform PCR amplification with a primer pair composed of F2 and R2 to obtain a PCR amplification product.
[0057] F2: 5'- GAAGAC TTAGGT ACAAACCTGCAAGATCAA-3';
[0058] R2: 5'- GGATCC TTA TGCTCCTGCAATCTTCAG-3'.
[0059] 3. The PCR amplified product of step 2 was double-digested with restriction endonucleases BbsI and BamHI, and the digested product was recovered.
[0060] 4. Digest the plasmid pHisSUMO with restriction endonucleases BbsI and BamHI to recover a vector backbone of about 5700 bp.
[0061] 5. Ligate the digested product of step 3 with the vector backbone of step 4 to obtain a recombinant plasmid. In the recombinant plasmid, the coding sequence of the VP2 protein coding...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com