A performance-improved recombinant Staphylococcus aureus protein a affinity ligand and its construction method
A staphylococcal protein and recombinant protein technology, applied in the field of genetic engineering, can solve the problems of difference in binding ability, different elution conditions, and inability to use strong alkali for a long time, and achieve the effect of good binding ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
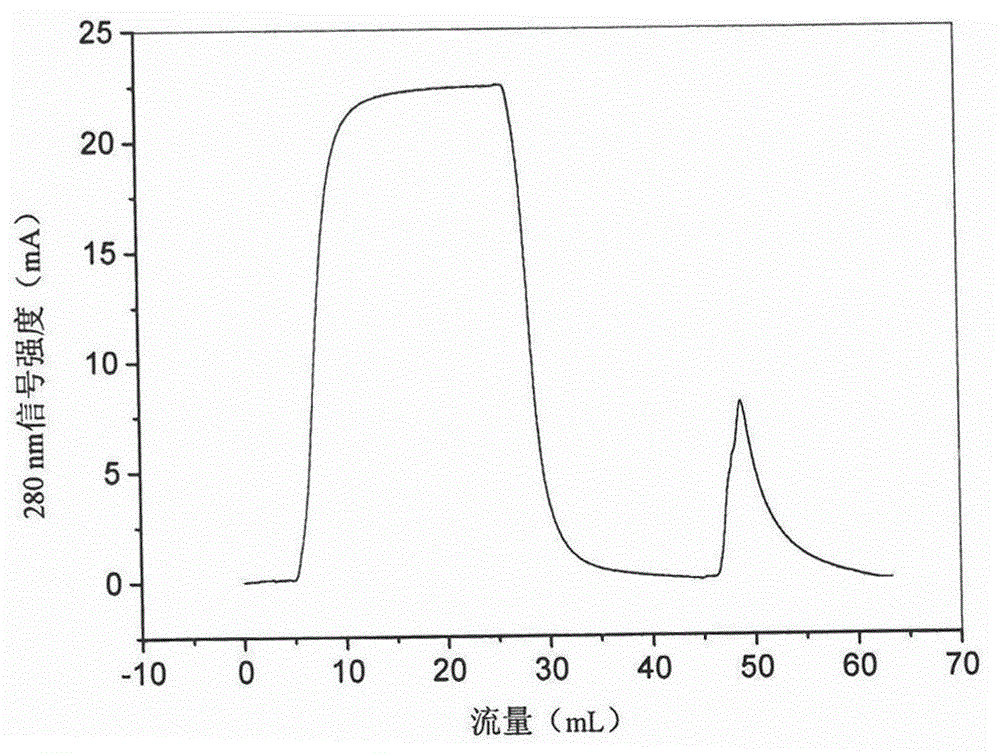
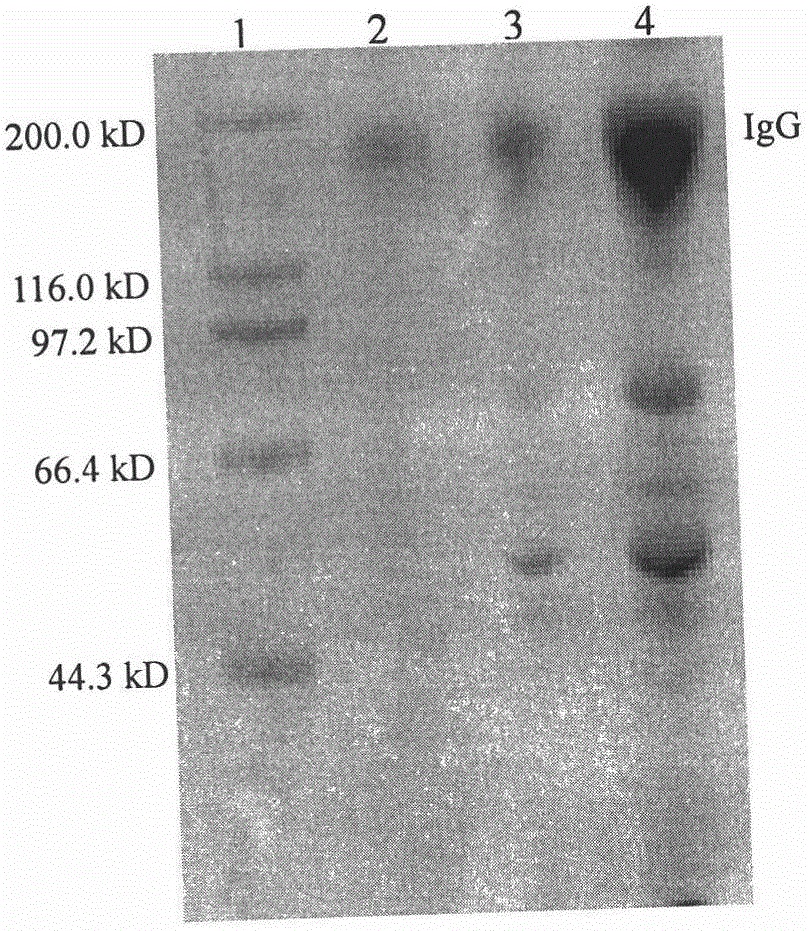
Image
Examples
Embodiment 1
[0025] The B segment of protein A was modified by overlapping PCR technology, and the modified B segment was named Z segment, and its expressed gene was z. According to the gene sequence provided by GeneBank, use the molecular biology software DNAMAN to design the forward primer F of z sequence 0 and reverse primer R 0 :
[0026] f 0 : CGGATAACAAATTCAAC
[0027] R 0 : TTA GCAACA TTTTGGTGCTTGTGC.
[0028] The forward primer and the reverse primer introduced Noc I and BamH I restriction sites, respectively, where the italic part is the restriction site, and the underlined part is the codon of the two cysteines added.
[0029] Add 6 glycines behind the second loop of the B fragment to reduce the binding force between protein A and IgG, expressed as Z(6G); in order to increase the alkali resistance of protein A, the 23rd asparagine and the 30th asparagine The phenylalanine at the position is replaced by threonine and alanine respectively, represented by Z(N23T, F30A)...
Embodiment 2
[0037] Extract Staphylococcus aureus DNA, utilize the F of embodiment 1 design 0 , R 0 As a primer, Staphylococcus aureus DNA as a template, using Pfu DNA polymerase to perform PCR on the B sequence. According to the instructions of Pfu DNA polymerase, select 50 μL to carry out.
[0038] PCR reaction conditions: pre-denaturation at 94°C for 5min; denaturation at 94°C for 40s, annealing temperature at 56°C for 1min30s, extension temperature at 72°C for 30s, and 30 cycles; finally, hold at 72°C for 10min.
[0039] Fragment B was recovered using an agarose gel recovery kit. Using the method of overlapping PCR, using Pfu DNA high-fidelity polymerase, using the recovered B fragment as a template, respectively using the F fragments designed in Example 1 0 and R 1 , F 1 and R 0 Perform PCR for the primers, and the reaction system and conditions are the same as above. The recovered products of PCR were mixed according to the volume ratio of 1:1, and the mixture was used as a te...
Embodiment 3
[0042] z n Indicates that n (n is 1-10) z sequences are connected end to end. Nhe I and BamHI were used as the first group of restriction endonucleases, and Xba I and BamH I were used as the second group of restriction endonucleases. Overnight culture of recombinant bacteria E.coli JM109 / pMD18-T-z obtained in Example 2 1 , plasmid extraction pMD18-T-z 1 , using two sets of enzymes to recombine the recombinant plasmid pMD18-T-z 1 Carry out enzyme cleavage reaction. The two groups of digested products were recovered from the gel, and the two groups of products were ligated overnight at 16°C using T4 DNA ligase to obtain a recombinant plasmid pMD18-T-z containing two z sequences connected end to end 2 , the pMD18-T-z 2 Transformed into competent E.coli JM109 for preservation.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com