Faded shewanella oneidensis LCT-SM262 strain in space environment
A technology of LCT-SM262, space environment, applied in the field of protein molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment 1
[0027] Specific implementation example 1, analysis of phenotypic characteristics of LCT-SM262:
[0028] 1. Morphological characteristics: Take 50ul sample for centrifugation, discard the supernatant, add 50ul of sterile water, absorb 20ul and fix it on the glass slide; for initial staining, absorb 2 drops of Gram stain I (crystal violet) on the glass slide sample Carry out initial dyeing for 1 minute, then rinse the dye solution with tap water; for mordant dyeing, absorb 2 drops of Gram stain II (iodine solution) to cover the painted surface and dye for about 1 minute, then rinse the dye solution with tap water, and absorb the water on the coated surface with absorbent paper; For decolorization, absorb 2 drops of Gram stain III (alcohol) on the coated surface for about 30 seconds, rinse the glass slide with water, and absorb the water on the coated surface with absorbent paper; counterstain, absorb Gram stain IV (safranin) Put 2 drops on the coated surface for about 1min, rins...
specific Embodiment 2
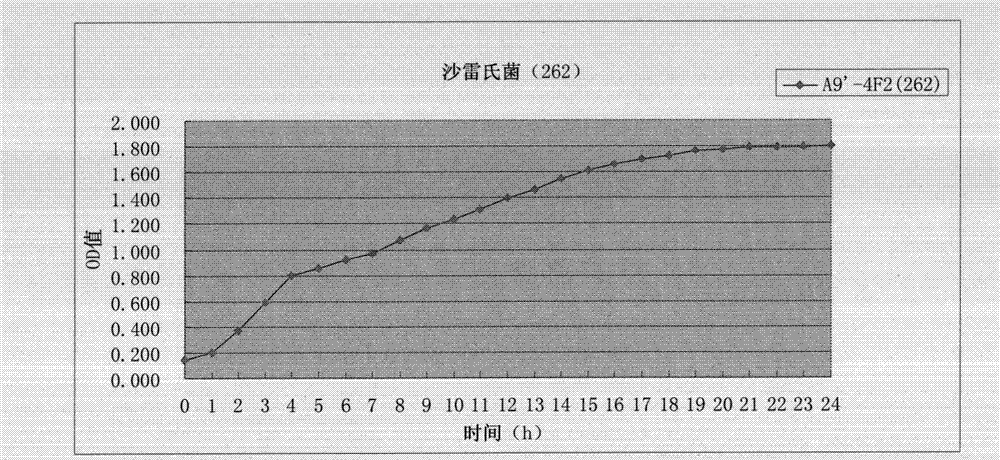
[0039] 6. Determination of growth curve: after activation, the strain was inoculated into a 100-hole honeycomb plate, and the growth curve was measured in the Bioscreen, measured for 24 hours, and the data was read. Calculate the average value of the obtained OD value, and then draw the curve, see image 3 . LCT-SM262 showed no difference in growth rate. Specific implementation example two, the genome and comparative genomics of LCT-SM262:
[0040] Cultivate Serratia discoloration and extract genomic DNA. First, use ultrasonic method to randomly interrupt the qualified DNA samples to obtain a series of DNA fragments. After treatment with T4 DNA polymerase, Klenow DNA polymerase and T4 polynucleotide kinase, etc. , recover the target fragment, obtain the sequencing library, and use Illumina’s Genome Analyzer II to sequence; the reads obtained after sequencing are filtered and then assembled and analyzed; Glimmer3.0 software is used to annotate genes and analyze SNPs, Indels, ...
specific Embodiment 3
[0045] Specific implementation example three, transcriptome sequencing analysis of LCT-SM262 strain:
[0046] After extracting the total RNA of the LCT-SM262 strain, the kit was used to remove the rRNA. Add fragmentation buffer to break mRNA into short fragments, use mRNA as a template, use six base random primers (random hexamers) to synthesize the first cDNA strand, then add buffer, dNTPs, RNase H and DNA polymerase I to synthesize the second strand The cDNA chain is purified by the QiaQuick PCR kit and eluted with EB buffer, then repaired at the end, added polyA and connected to the sequencing adapter, then used agarose gel electrophoresis to select the fragment size, and finally performed PCR amplification. The sequencing library was sequenced using Illumina HiSeq 2000. After removing the impurity data from the original sequence data, count rRNA statistics, evaluate the distribution of Reads on the genome, gene coverage, count gene expression differences, analyze differen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com