Construction method for pichia pastoris bacterial strain of high-yield S-ademetionine
A technology for adenosylmethionine and construction methods, applied in the direction of microorganism-based methods, botany equipment and methods, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
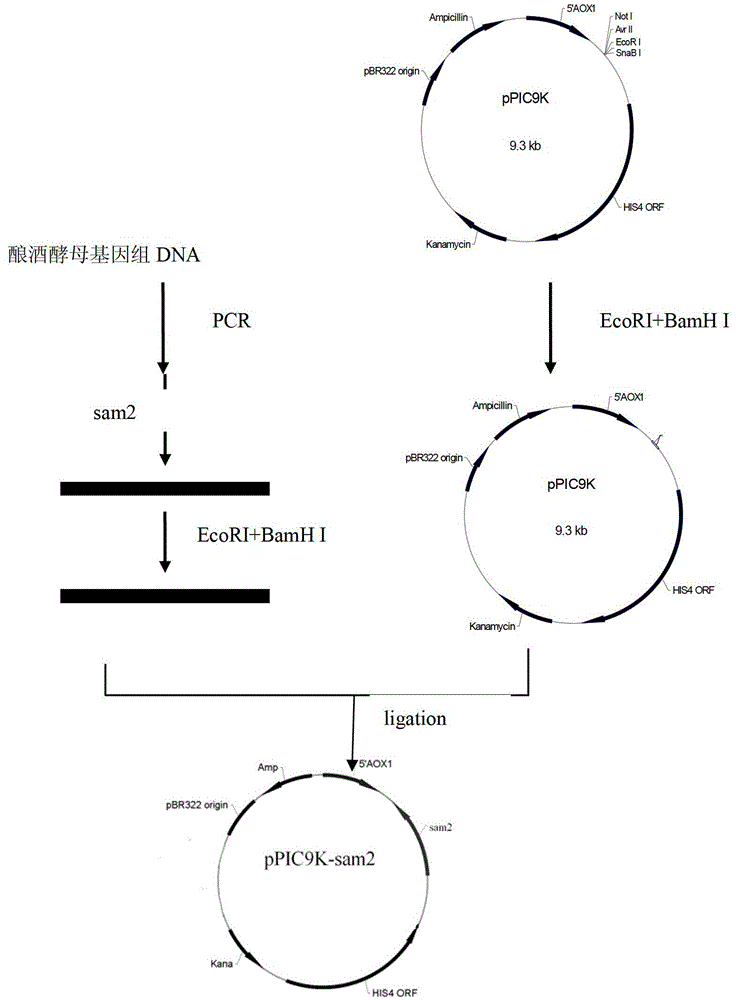
[0028] Example 1. Construction of recombinant expression vector pPIC9K-sam2
[0029] The gene sam2 was amplified from the S. cerevisiae genome using primers sam2-F: 5'-CAGGATCCACCATGACCAAGAGCAAAACT-3'; EcoR I and Bam H I respectively double-digested the PCR amplification product and pPIC9K plasmid DNA, recovered the target band by agarose gel electrophoresis, and used T 4 DNA ligase ligated the recovered target bands to obtain the eukaryotic expression vector pPIC9K-sam2, and used CaCl 2 Transform it into Escherichia coli DH5α by heat shock at 42°C for 90 seconds. Single colonies were screened for Kana resistance. The selected transformed clones were identified by PCR and enzyme digestion to prove that the clones were correct and then sent to Dalian Bao Biological Company for sequencing. For specific operations, see figure 1 .
Embodiment 2
[0030] Example 2. Obtaining of cbs gene knockout plasmid T-C-Z and fragment C-Z
[0031] Using primers cbs-F: 5'-TTCTGGAGCACATTGGAA-3'; cbs-R: 5'-AGTGTATGCCTAG ATGG-3', the cbs gene was amplified from the Pichia pastoris genome, and then subcloned into the vector pMD19T to obtain a recombinant vector T-cbs. Using primers zeocin-F: 5'-GGACTAGTAGACCTTCGTTTGTGC-3'; zeocin-R: 5'-GGACTAGTCGGTTCCTGGCCTTTTG-3' to amplify the zeocin gene from pPICZα-A, use Speech I digest the PCR product zeocin gene and the recombinant plasmid T-cbs respectively, and use T 4 DNA ligase ligated the recovered fragments to obtain the cbs gene knockout plasmid T-C-Z. After successful identification by PCR and enzyme digestion, it was sent to Dalian Bao Biological Company for sequencing. After successful sequencing, use Sal I and Bgl II double-digested the recombinant plasmid T-C-Z, recovered a 2100bp fragment from rubber tapping, and named it C-Z. See the specific operation process figure...
Embodiment 3
[0032] Example 3. Transformation and screening of recombinant Pichia pastoris
[0033] The identified correct recombinant expression vector pPIC9K-sam2 plasmid DNA was subjected to endonuclease Bpu1102I linearization treatment, electroporation transformation of Pichia pastoris GS115. Immediately after the electric shock, 1ml of pre-cooled 1mol / l sorbitol was added, centrifuged at 3000rpm for 5min, the bacteria were resuspended in 400μl of pre-cooled 1mol / l sorbitol, and 200μl was spread on MD plates (1.34% YNB, 4×10 -5 % biotin, 2% glucose, 2% agarose) at 30°C until colonies appeared. Randomly pick colonies and inoculate them on YPD plates containing different concentrations of -418 (0, 0.50mg / ml, 1.00mg / ml, 2.00mg / ml, 3.00mg / ml, 4.00mg / ml), and culture them at 30°C for 2-5 days. , and check the colony growth every day. The positive clone transformant ZJGSU01 with high resistance to -418 was quickly screened out according to the growth conditions.
[0034] Transform the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com