Primers, probes, a test kit and a test method for triple real-time fluorescence PCR detection of four bacteria
A real-time fluorescence, primer detection technology, applied in biochemical equipment and methods, fluorescence/phosphorescence, microbial determination/inspection, etc. System amplification efficiency evaluation and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] The selection of embodiment 1 strain and the design of detection primers and detection probes
[0093] A total of 142 strains of 34 different serotypes of Salmonella were selected, including 1 standard strain of Salmonella paratyphi C (IQCC10527), 21 isolates of Salmonella paratyphi C (SP1-SP21), and 1 standard strain of Salmonella choleraesuis (IQCC10502). , 24 isolates of Salmonella choleraesuis (SC1-SC24), 1 standard strain of Salmonella typhi (CMCC50071), 30 isolates of Salmonella typhi (ST1-ST30), 1 standard strain of Salmonella gallinarum typhi (CMCC50770), Salmonella typhi 33 strains (SG1-SG33) and 30 strains of other serotypes of Salmonella were isolated. In addition, 21 strains of non-Salmonella such as Proteus were selected. The strain information is shown in Table 1. The above strains were confirmed by API20E reagent strips and serological tests. The above-mentioned strains come from the China General Microorganism Culture Collection Management Center, the G...
Embodiment 2
[0122] Embodiment 2: Preparation of template DNA
[0123] The 22 strains of Salmonella paratyphi C, 25 strains of Salmonella choleraesuis, 31 strains of Salmonella typhi, 34 strains of Salmonella gallinarum typhi, 30 strains of other serotypes of Salmonella and 21 strains of non-Salmonella were cultured in buffered peptone water (BP) at 37°C for 10 hours , then take 10ml of the above-mentioned cultured bacteria solution and inoculate them into 90ml of TTB enrichment solution, incubate at 44.5°C for 18 hours, then take 1ml of the above-mentioned cultured bacteria suspension and transfer them into centrifuge tubes, centrifuge at 12000r / min for 5min to remove the supernatant, and use 1ml of deionized The water floats and precipitates, and then centrifuges at 12000r / min for 3min to remove the supernatant, repeats twice, and finally adds 200μl deionized water, and extracts DNA on a nucleic acid extractor for real-time fluorescent PCR amplification.
Embodiment 3
[0124] Embodiment 3: detection primer and detection probe specificity test
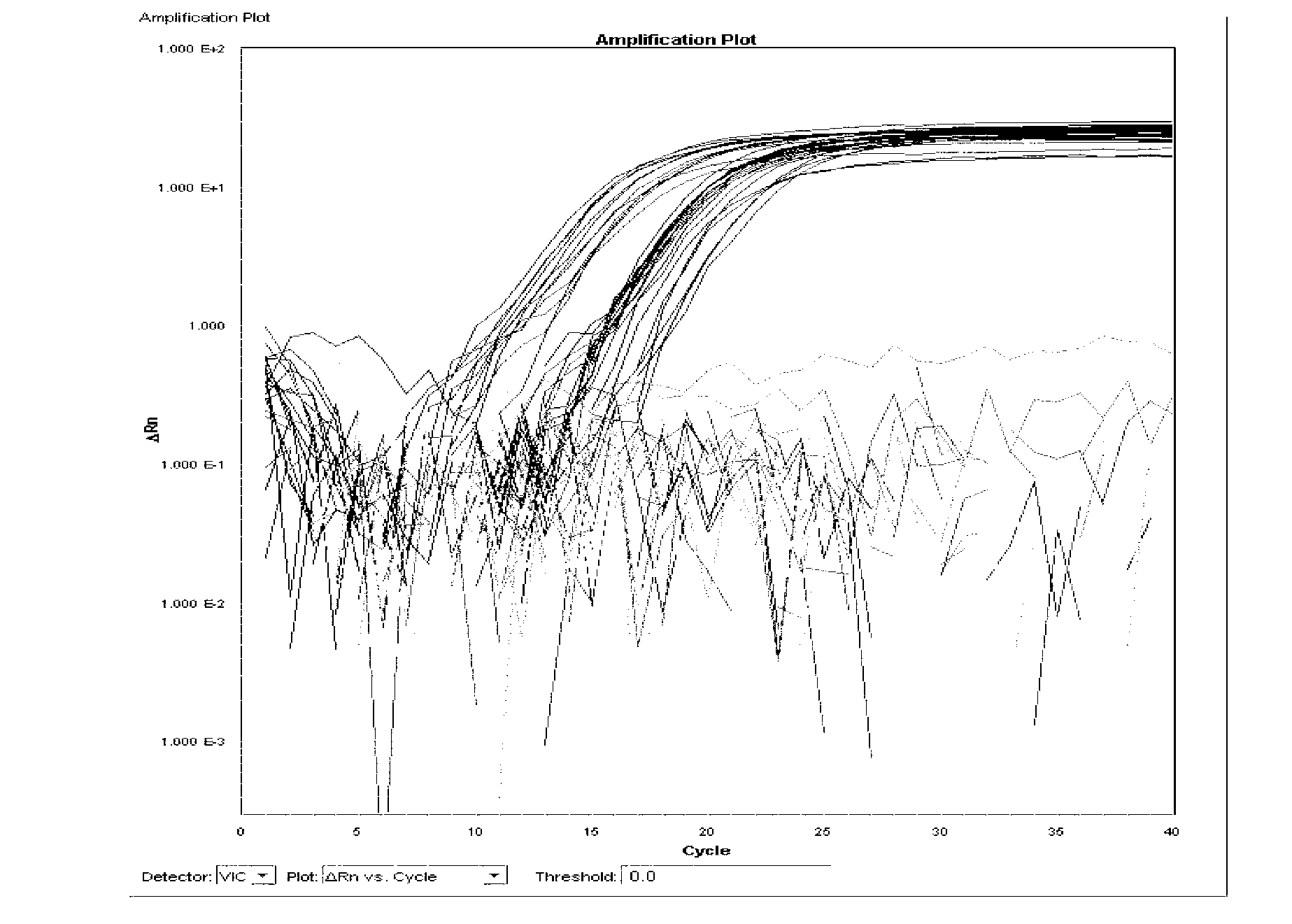
[0125] Single real-time fluorescent PCR reaction system 30μl, including: template DNA 1μl, 10×TaqMan buffer 4μl, 5mmol / LMgCl 2 2 μl, 2.5 mmol / L dNTPs 3 μl, 20 μmol / l detection probe 1 μl, 20 μmol / l detection primers 1 μl (total 2 μl), 0.55U UNG enzyme 0.2 μl, 2.5U / μl Taq polymerase 3 μl, deionized water 13.8 μl. The fluorescent PCR reaction parameters were 95°C for 30s, 95°C for 5s, 60°C for 34s, and 40 cycles.
[0126] 1. Specificity test 1: Specificity test of Salmonella choleraesuis and Salmonella paratyphi C
[0127] Extract the DNA of 25 strains of Salmonella choleraesuis, 22 strains of Salmonella paratyphi C, 31 strains of Salmonella typhi, 34 strains of Salmonella gallinarum typhi, 30 strains of other serotype Salmonella and 21 strains of non-Salmonella according to the method described in Example 2, according to the above A single real-time fluorescent PCR reaction system is used for real-ti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com