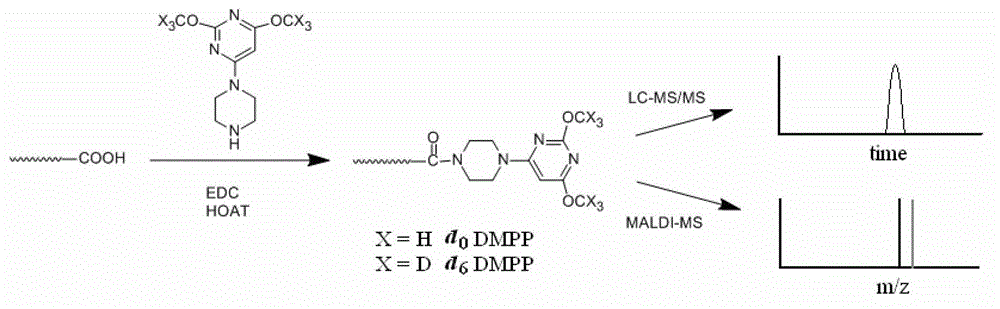
Use of piperazinopyrimidine isotope labeling reagent
A technology of isotope labeling and base pyrimidine, which is used in measurement devices, material analysis by electromagnetic means, instruments, etc., can solve the problems of weak mass spectral response, difficult qualitative and quantitative detection, weak content, etc., to avoid signal interference and improve product quality. The effect of mass spectral response intensity and product background noise is low
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: [d 0 ]- or [d 6 ]-2,4-dimethoxy-6-piperazinyl pyrimidine, (abbreviated as [d 0 ]- / [d 6 ]-DMPP) synthesis.
[0045] The operation steps are as follows:
[0046] Boc-protected piperazine (373mg, 2mmol) and potassium carbonate (304mg, 2.2mmol) were dissolved in an appropriate amount of 20ml of anhydrous DMF, and 2,4,6-trichloropyrimidine (0.23ml ,2mmol), and stirred gently. After reacting at room temperature for 10 hours, the reaction was quenched by pouring into water, and extracted with dichloromethane to obtain crude product 2,4-dichloro-6-Boc-piperazinylpyrimidine (466mg, 1.4mmol). Dissolve 150mg of sodium metal in 15mL of anhydrous methanol to form a sodium methoxide solution. Dissolve 2,4-dichloro-6-Boc-piperazinylpyrimidine in 10 mL of anhydrous methanol, then gradually add it dropwise into the prepared sodium methoxide solution, stir overnight, and place the reaction solution at 25°C and stir overnight, Then it was concentrated by filtration, then...
Embodiment 2
[0066] Embodiment 2: comparative analysis of polypeptide
[0067] The operation steps are as follows:
[0068] 1. Place bovine serum albumin in an aqueous solution containing 0.04-0.05M ammonium bicarbonate and pH 8.0-8.5 for heat denaturation for 10 minutes.
[0069] 2. After cooling to room temperature, add trypsin to the solution at a weight ratio of 50:1, and enzymolyze it at 37°C for 12-16 hours.
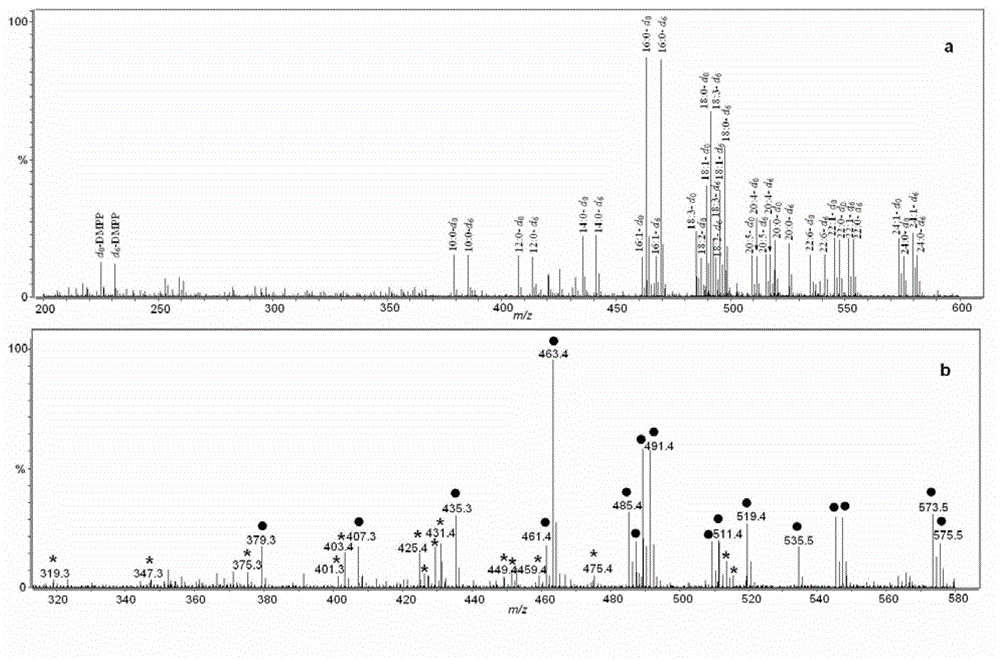
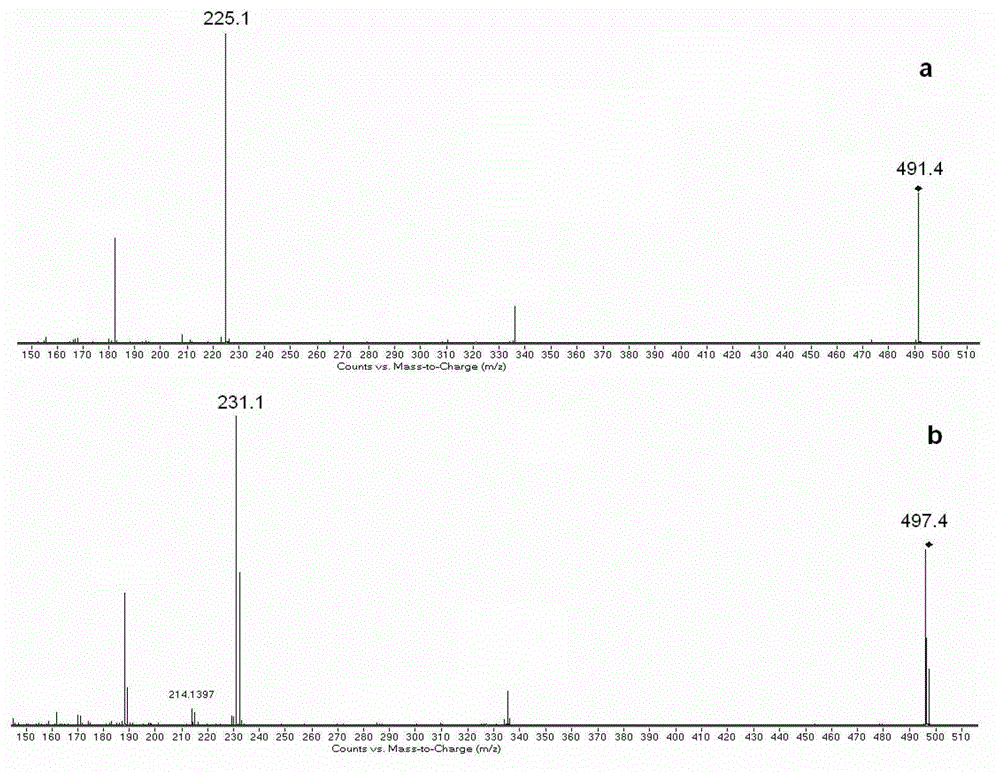
[0070]3. Divide the enzymatic hydrolysis product and standard polypeptide a and n into two parts, and react the two parts with the light and heavy labeling reagent DMPP respectively, the solvent is acetonitrile, perform the same post-processing steps as in Example 1, but without resin adsorption, etc. Subsequent steps, followed directly by mass spectrometry.
[0071] 4. The analysis conditions of MALDI-TOF MS are: all the acquisition conditions are carried out in the positive ion mode, and the sample matrix CHCA solvent is 0.1% trifluoroacetic acid and 50% acetonitrile water....
Embodiment 3
[0078] Example 3: Labeling of proteins
[0079] The operation steps are as follows:
[0080] 1. As in Example 1, undenatured proteins (lysozyme, bovine serum albumin, BSA) were reacted with light labeling reagents, and the same post-processing steps were performed.
[0081] 2. Mix the unlabeled protein with the labeled protein at the same concentration.
[0082] 3. The analysis conditions of MALDI-TOF MS are: all acquisition conditions are carried out in the positive ion mode, the laser energy is appropriately increased by 20%, the detection mode is linear, and the spotting matrix SA solvent is 50% of 0.1% trifluoroacetic acid Acetonitrile in water.
[0083] The analysis results of MALDI-TOF MS are as follows:
[0084] 1. If Figure 9 As shown, both bovine serum albumin and lysozyme are completely labeled, and compared with the unlabeled original protein, the peak area is increased by more than five times, which is very beneficial for the detection of low kurtosis protein....
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com