Rapid screening method of zinc finger protein
A zinc finger protein and screening method technology, applied in the biological field, can solve problems such as heavy workload, low success rate, and difficult implementation, and achieve the effect of cost reduction and reliable screening results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Taking the screening of zinc finger proteins targeting human PGRN as an example to construct a zinc finger nuclease targeting human PGRN gene, the steps are as follows:
[0040] 1. Construction of a small zinc finger protein library targeting human PGRN
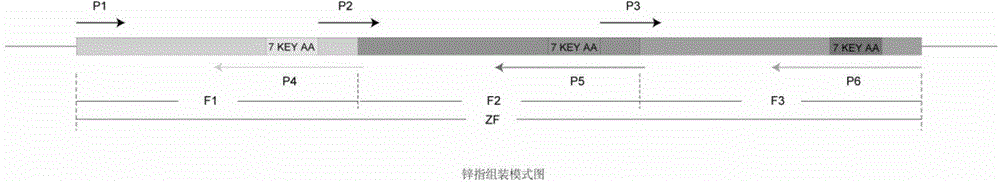
[0041] Since zinc finger nucleases exist in pairs and form dimers to play a role, it is necessary to screen a pair of zinc finger proteins. The binding sites of the two zinc finger proteins are respectively located on the two complementary DNA strands of the target gene. The recognition sites are separated by 5-7 bases, so the final target sequence is a 23-25bp DNA fragment. ZiFiT, an online zinc finger protein prediction software provided by the Zinc Finger Consortium, provides a service dedicated to predicting paired zinc finger protein target sites for zinc finger nucleases. The specific method is as follows:
[0042] Find the genome sequence NG_007886 of human PGRN from NCBI, and use the online zinc finger protei...
Embodiment 2
[0142] Taking the screening of zinc finger protein targeting human PGRN as an example, the steps are as follows:
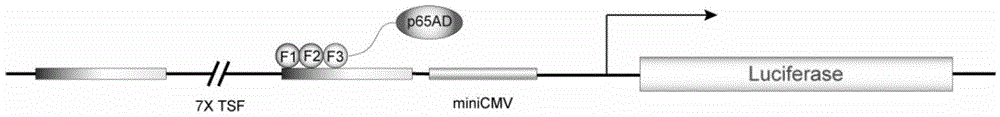
[0143] In the synthesis step 2-(1) of the nuclear localization signal and detection label in Example 1, different detection labels are added to the 5' end of the nuclear localization signal of the chimeric transcription factor assembled from the left and right zinc finger proteins, They are the His tag on the left, the myc tag on the right, and the detection tags on the left and right can also be replaced with each other. The left and right nuclear localization signals and detection tags were synthesized in the full length of Shanghai Sangong, and their sequences are:
[0144] left side
[0145] ATCGATATGCACCATCACCACCATCACGCACCAAAGAAAAAGCGGAAGGTAGATTCAAAGATCATGATGGCGATTACAAGGACCACGATATCAAGCTT
[0146] Right
[0147]GAATTCATGCAGAAGCTGATCAGCGAGGAGGACCTGGCGCCCAAGAAGAAACGAAAGGTCTATCTCTTACGATGTGCCAGACTACGCCGGGTATCCATATGATGTGCCTGACTATGCCGGCAGCGAGGGTACC
[0148] In t...
Embodiment 3
[0163] In the nuclear localization signal synthesis step 2-(1) of Example 1, the full-length nuclear localization signal was synthesized in Shanghai Sangon, and its sequence is:
[0164] left side:
[0165] ATcGAtATGGCACCAAAGAAAAAGCGGAAGGTAGATTACAAGATCATGATGGCGATTACAAGGACCACGATATCAAGCTT;
[0166] Right:
[0167] GAATTCATGGCGCCCAAGAAGAAACGAAAGGTCTATCCTTACGATGTGCCAGACTACGCCGGGTATCCATATGATGTGCCTGACTATGCCGGAGCGGTACC
[0168] At the same time, ClaI and HindIII sites were introduced at both ends of the left nuclear localization signal, and EcoRI and KpnI sites were introduced at both ends of the right nuclear localization signal.
[0169] In the cloning step 2-(2) of the transcriptional activation domain p65AD of the transcription factor p65, the following primers were designed and synthesized according to the DNA sequence of p65AD, because different enzyme cleavage sites were introduced at the two ends of the zinc finger protein on the left and right point, respectively cloned p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com