Triple real-time fluorescent PCR detection primers, probes, detection kits and detection methods for three bacteria
A technology for real-time fluorescence and detection probes, applied in biochemical equipment and methods, microbial-based methods, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] A total of 15 strains of Salmonella enteritidis were selected, of which 1 strain was the standard strain ATCC13076, 13 strains were isolated from human sources, and 1 strain was isolated from frozen chicken samples; a total of 11 strains of Salmonella typhimurium were selected, of which 1 strain was the standard strain ATCC14028, 10 The strains were food isolates (as shown in Table 1); a total of 21 strains of different serotypes of non-Salmonella enteritidis and non-typhimurium isolated from food were selected (as shown in Table 2); 1 strain of Salmonella Dublin CMCC 50042 was selected; There were 11 negative control strains, including Listeria monocytogenes ATCC19115, Staphylococcus aureus ATCC6538, Enterobacter sakazakii ATCC29544, Escherichia coli ATCC25922, Shigella baumannii CMCC51582, Shigella sonnei CMCC51334, dysentery Shigella NICPBP51252, Yersinia enterocolitica CMCC52221, Yersinia pseudotuberculosis CMCC53510, Klebsiella pneumoniae CMCC46102, Vibrio parahaemo...
Embodiment 2
[0096]The 21 strains of non-Salmonella enteritidis non-typhimurium different serotype Salmonella strains (shown in Table 2), 15 strains of Salmonella enteritidis, 11 strains of Salmonella typhimurium, 1 strain of Salmonella Dublin CMCC 50042 and 11 negative control strains were used Buffered peptone water (BP) was cultured at 37°C for 10 hours, then 1ml of the cultured bacteria was inoculated into the TTB enrichment solution, cultured at 44.5°C for 18 hours, and then 1ml of the cultured bacteria suspension was transferred into a centrifuge tube, 12000r / min Centrifuge for 8 minutes to remove the supernatant, float the sediment with 1ml of deionized water, then centrifuge at 12000r / min for 5 minutes to remove the supernatant, repeat twice, finally add 200μl of deionized water, extract DNA on a nucleic acid extractor for real-time fluorescent PCR amplification.
[0097] Embodiment 3: detection primer and detection probe specificity test
Embodiment 3
[0098] Single-plex real-time fluorescent PCR reaction system 30μl, including: template DNA 1μl, 10×TaqMan buffer 4μl, 5mmol / L MgCl 2 2μl, 2.5mmol / L dNTPs 3μl, 20μmol / l TaqMan detection probe 1μl, 20μmol / l detection primer 1μl each (total 2μl), 0.55U UNG enzyme 0.2μl, 2.5U / μl Taq polymerase 3μl, deionized water 13.8 μl. The parameters of the fluorescent PCR reaction were 95°C for 30s, 95°C for 5s, 60°C for 34s, and 40 cycles.
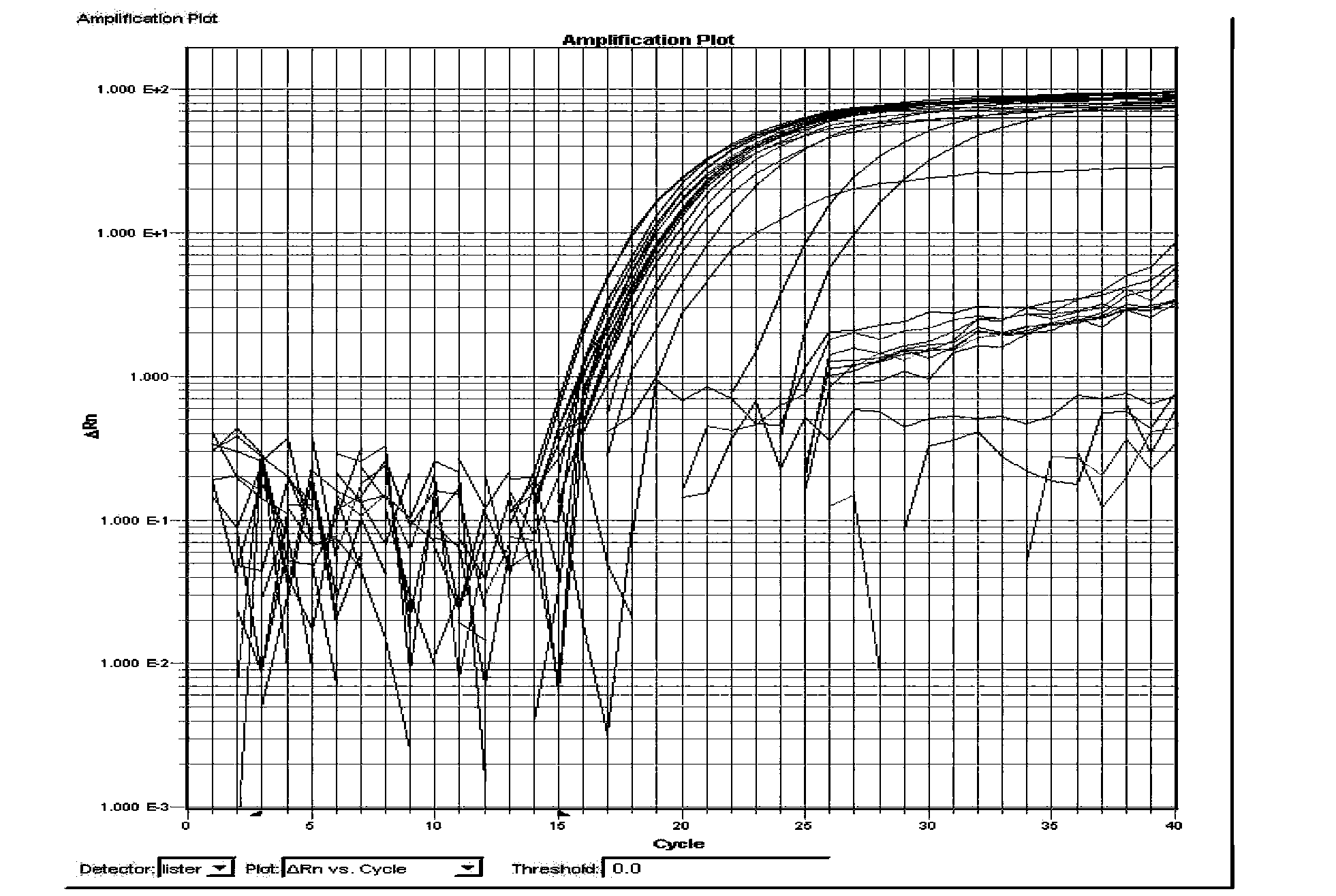
[0099] 1. Specificity test 1: Specificity test of detection primers and detection probes for Salmonella
[0100] According to the method described in Example 2, 21 different serotype Salmonella strains of non-Salmonella enteritidis non-Typhimurium, Salmonella typhimurium standard strain ATCC14028, Salmonella enteritidis standard strain ATCC 13076, Salmonella Dublin CMCC 50042 and 11 negative control strains were extracted. DNA is amplified by real-time fluorescent PCR according to the above-mentioned single-plex real-time fluorescent PCR reaction syste...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com