Glucoamylase producing method
A technology of glucoamylase and gene expression, which is applied in the field of constructing microbial engineering bacteria to produce recombinant protein and producing recombinant glucoamylase, which can solve the problem of narrow reaction temperature range and suitable reaction pH range, and cannot satisfy the reaction temperature and suitable reaction pH range Demand and other issues to achieve the effect of saving raw materials
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
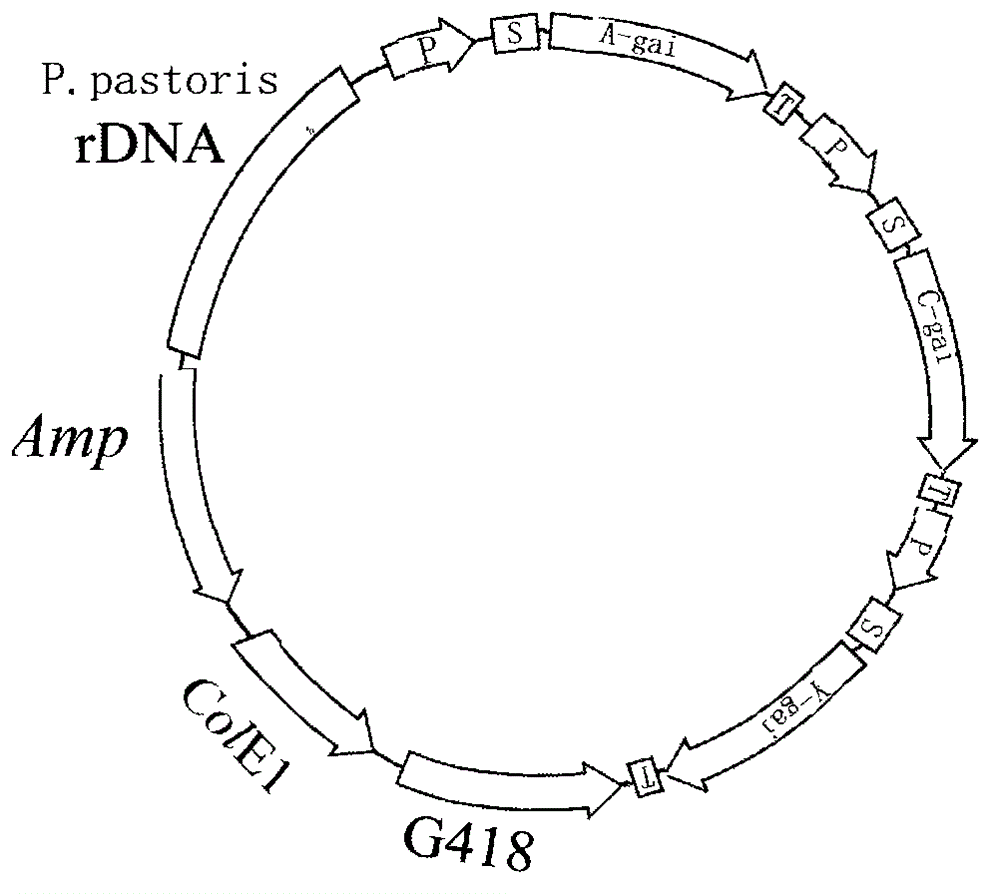
[0022] 1.1 Pichia pastoris expression vector containing the glucoamylase gene expression framework of Aspergillus, the glucoamylase gene expression framework of Chaetomium and the glucoamylase gene expression framework of yeast
[0023] 1.1.1 Construction of cloning vector
[0024] A professional DNA sequence synthesis company synthesizes two base complementary double strands containing ampicillin (AMP) gene sequence, polyclonal adapter and E. coli replication origin, and forms cohesive ends at both ends of each DNA strand sequence. It is circularized by the action of DNA ligase to form a DNA cloning vector. The cloning vector was named pPA.
[0025] 1.1.2 Acquiring genes
[0026] ① Amplification of Aspergillus glucoamylase gene by reverse transcription PCR
[0027] Primer 1: 5'CT GGATCC atgtcgttccgatctcttct3'[Description: The 8 bases of 5' are the restriction enzyme protection bases (2 bases) and the enzyme recognition site (6 underlined bases)]
[0028] Primer 2: 5'aa ...
Embodiment 2
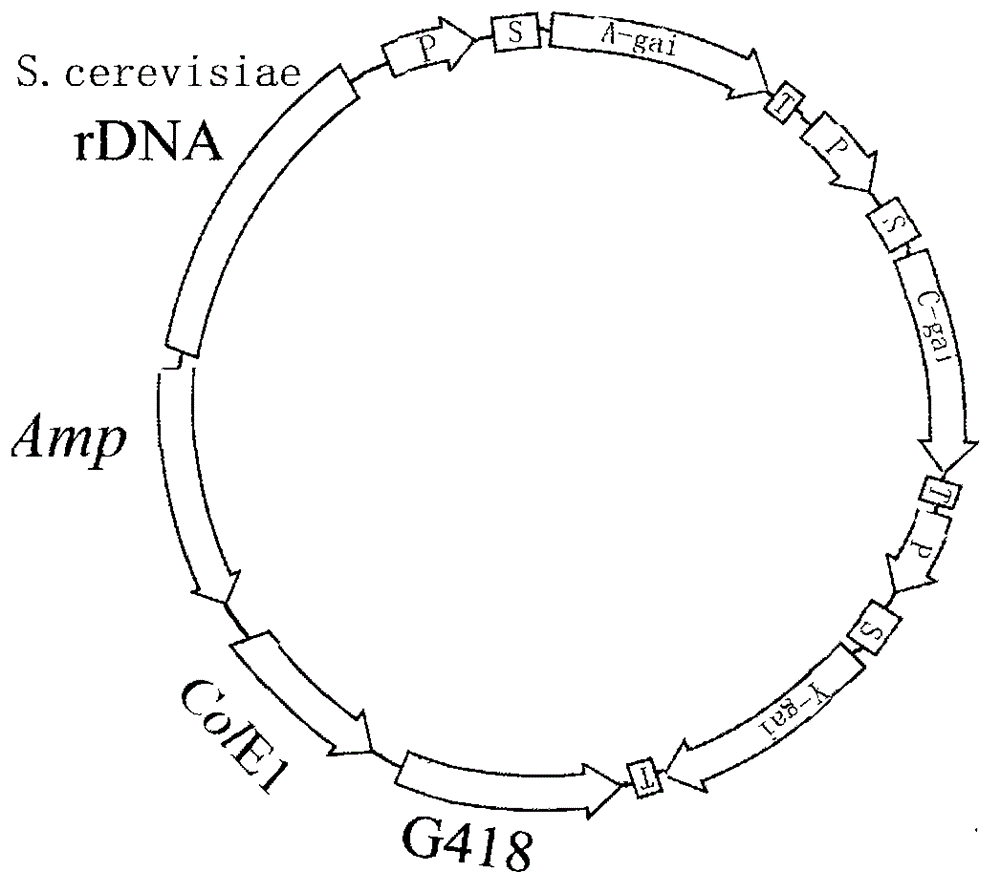
[0075] 2.1 Saccharomyces cerevisiae expression vector containing the glucoamylase gene expression framework of Aspergillus, the glucoamylase gene expression framework of Chaetomium and the glucoamylase gene expression framework of yeast
[0076] 2.1.1 Construction of cloning vector
[0077] A professional DNA sequence synthesis company synthesizes two base complementary double strands containing ampicillin (AMP) gene sequence, polyclonal adapter and E. coli replication origin, and forms cohesive ends at both ends of each DNA strand sequence. It is circularized by the action of DNA ligase to form a DNA cloning vector. The cloning vector was named pSA.
[0078] 2.1.2 Acquiring genes
[0079] ④ Amplification of Aspergillus glucoamylase gene by reverse transcription PCR
[0080] Primer 1: 5'CT GGATCC atgtcgttccgatctcttct3'[Description: The 8 bases of 5' are the restriction enzyme protection bases (2 bases) and the enzyme recognition site (6 underlined bases)]
[0081] Primer...
Embodiment 3
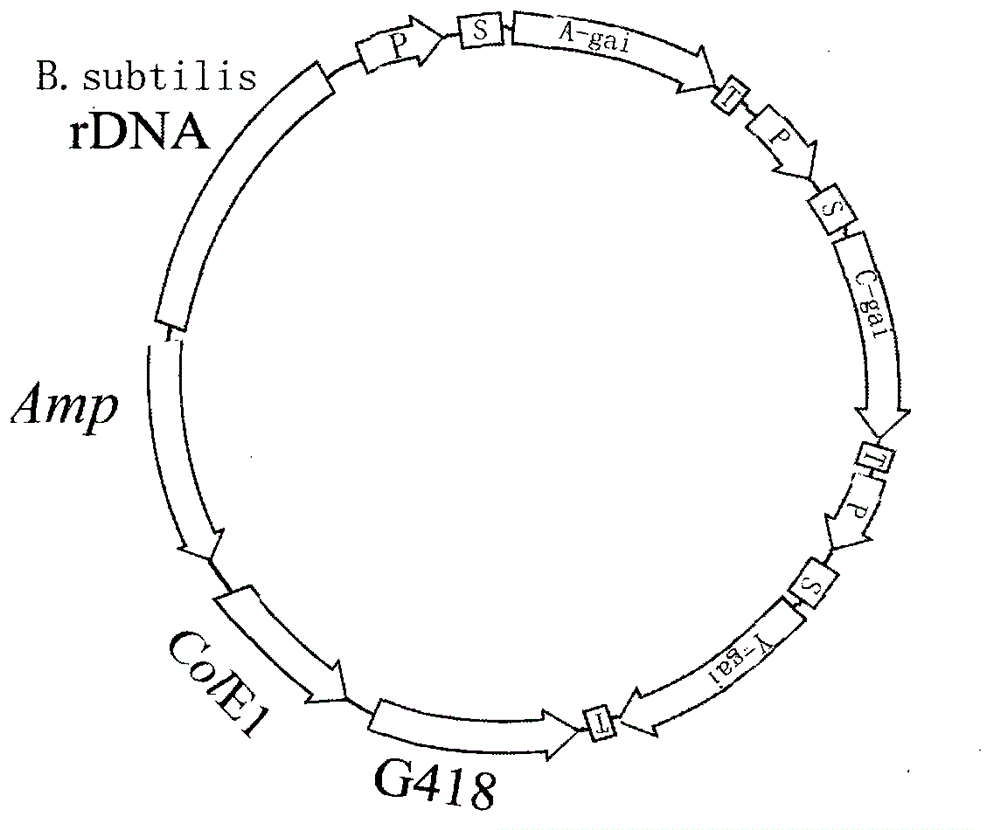
[0134] 3.1 Bacillus subtilis expression vector containing the glucoamylase gene expression framework of Aspergillus, the glucoamylase gene expression framework of Chaetomium and the glucoamylase gene expression framework of yeast
[0135] 3.1.1 Construction of cloning vector
[0136] A professional DNA sequence synthesis company synthesizes two base complementary double strands containing ampicillin (AMP) gene sequence, polyclonal adapter and E. coli replication origin, and forms cohesive ends at both ends of each DNA strand sequence. It is circularized by the action of DNA ligase to form a DNA cloning vector. The cloning vector was named pBA.
[0137] 3.1.2 Acquiring genes
[0138] Amplification of Aspergillus Glucoamylase Gene by Reverse Transcription PCR
[0139] Primer 1: 5'CT GGATCC atgtcgttccgatctcttct3'[Description: The 8 bases of 5' are the restriction enzyme protection bases (2 bases) and the enzyme recognition site (6 underlined bases)]
[0140] Primer 2: 5'aa ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com