Functional specific molecular marker PikFNP for resistance genes Pik of rice blast and method and application of functional specific molecular marker PikFNP
A resistance gene and molecular marker technology, applied in the field of agricultural biology, can solve the problems of error in identification results and low selection efficiency of target genes, and achieve the effect of reducing labor costs and improving the efficiency of breeding work.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] Example 1: Alignment of Pik allele coding region sequence and SNP identification
[0040] Amplification by conventional PCR (Yuan et al. 2011. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NB S-LRR genes. Theor Appl Genet 122: 1017-1028; Zhai et al. al.2011.The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication.New Phytologist189:321-334), sequencing (Shanghai Yingjun Biotechnology Co., Ltd., Guangzhou Branch), from 5 disease resistance Rice variety Shin 2 (Pik-s donor variety; Kiyosawa.1987. With genetic view on the mechanism of resistance and virulence. Japanese Journal of Genetics 41: 89-92), Tsuyuake (Pik-m donor variety; Ashikawa et al .2008.Two adjacent nucleotide-binding site-leucine rich repeat class genes are required to confer Pikm-specific rice blast resistance.Genetics180: 2267-2276), Kusabue (Pik donor variety; Zhai et al.2011. The isolation and characteriza...
Embodiment 2
[0042] Example 2: Pik functional specific molecular marker and its primer design and detection
[0043] According to the principle of dCAPS labeling (Neff et al., 2002. Web-based primer design for the single nucleotide polymorphism analysis. Trends in Genetics 18: 613-615), the functional specificity is designed at 100-200bp upstream of the above SNP site The upstream primer Pik-FNP-F is shown in SEQ ID NO.1; the functionally specific downstream primer Pik-FNP-R with mismatched bases is designed at the above SNP site, as shown in SEQ ID NO.2 . Use this set of primers to amplify the DNA template of the variety carrying the Pik gene and other alleles.
[0044] The PCR amplification reaction system of Pik-FNP is as follows:
[0045]
[0046] Pik-FNP PCR amplification reaction temperature cycle conditions are as follows: 94°C for 3 minutes; 94°C for 30 seconds, 58°C for 30 seconds, 72°C for 1 minute, 35 cycles; 72°C for 5 minutes; 10°C storage.
[0047] After the PCR reaction is complet...
Embodiment 3
[0050] Example 3: Application of specific molecular markers of resistance gene Pik in identifying different rice blast resistance genes
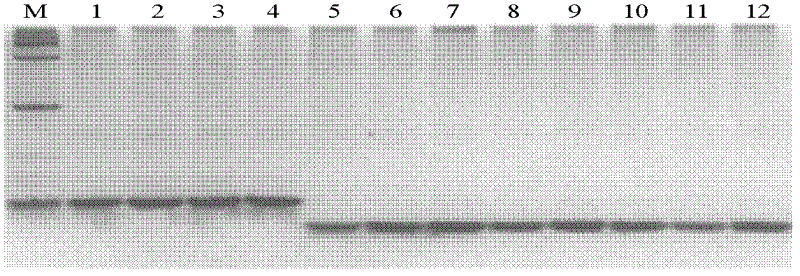
[0051] Extracted 12 rice varieties containing different rice blast resistance genes [respectively: Kusabue, Kando51, Kuyuku 131, GA20, Taifenzhan (Zhai et al.2011. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication.New Phytologist 189:321-334), Tsuyuake(Ashikawa et al.2008.Two adjacent nucleotide-binding site-leucine rich repeat class genes are required to confer Pikm-specific rice blast resistance.Genetics 180:2267-2276) , K60(Yuan et al.2011.The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes.Theor Appl Genet 122: 1017-1028), IRBLk-Ka, IRBLKm- Genomic DNA of TS, IRBLkp-K60, IRBLks-S, and IRBLKh-K3 (Kobayashi et al. 2007. Development of new sets of international standard differential varieties for blast resistance in rice (Oryza ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com